Normal Mode Analysis
nma.RdPerform normal mode analysis (NMA) on either a single or an ensemble of protein structures.
nma(...)
Arguments
| ... | arguments passed to the methods
For function For function |
|---|
Details
Normal mode analysis (NMA) is a computational approach for studying and characterizing protein flexibility. Current functionality entails normal modes calculation on either a single protein structure or an ensemble of aligned protein structures.
This generic nma function calls the corresponding
methods for the actual calculation, which is determined by the class
of the input argument:
Function nma.pdb will be used when the input argument is
of class pdb. The function calculates the normal modes of a
C-alpha model of a protein structure.
Function nma.pdbs will be used when the input argument is
of class pdbs. The function will perform normal mode analysis
of each PDB structure stored in the pdbs object
(‘ensemble NMA’).
See documentation and examples for each corresponding function for more details.
References
Skjaerven, L. et al. (2014) BMC Bioinformatics 15, 399. Grant, B.J. et al. (2006) Bioinformatics 22, 2695--2696.
Author
Lars Skjaerven
See also
Examples
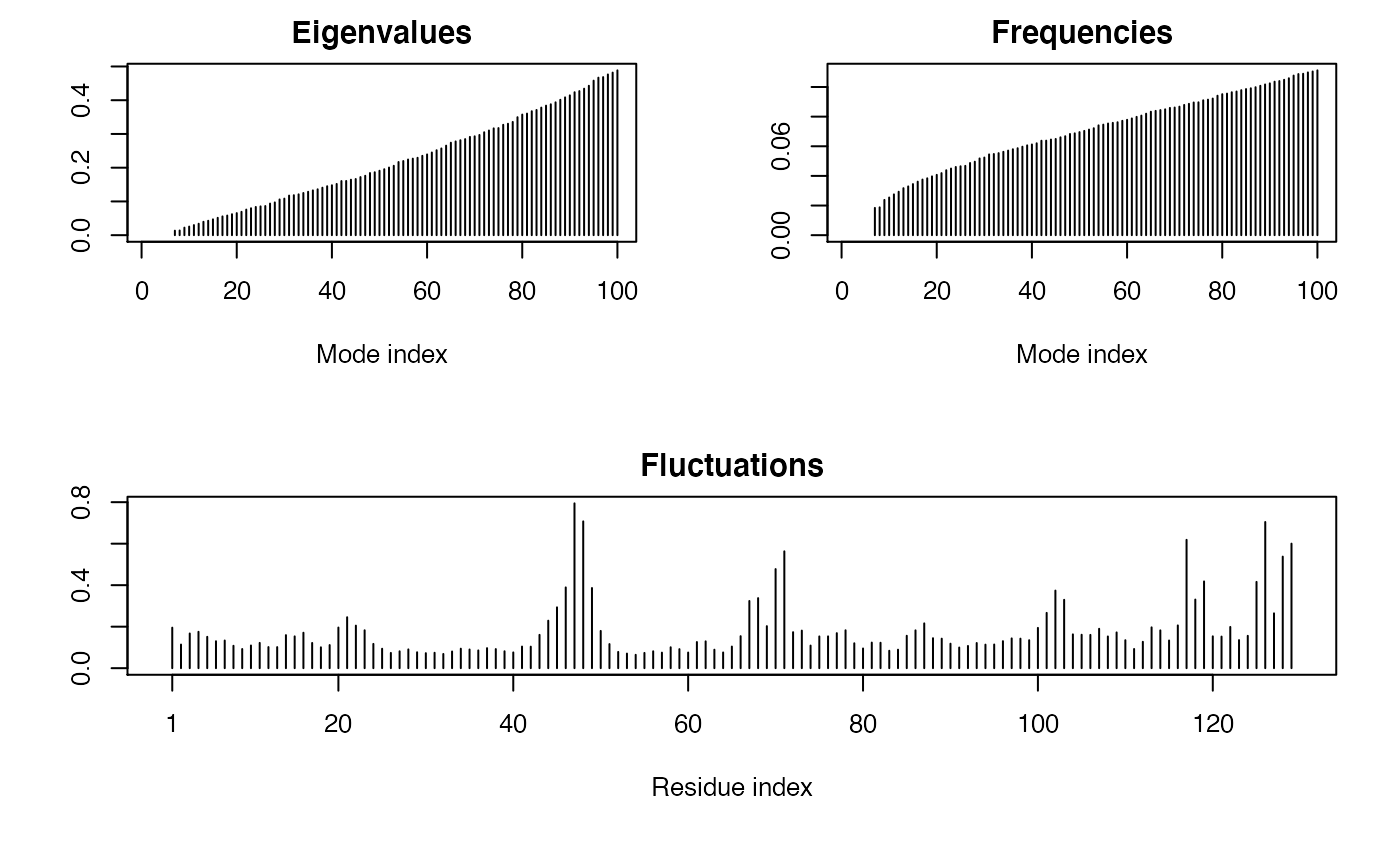
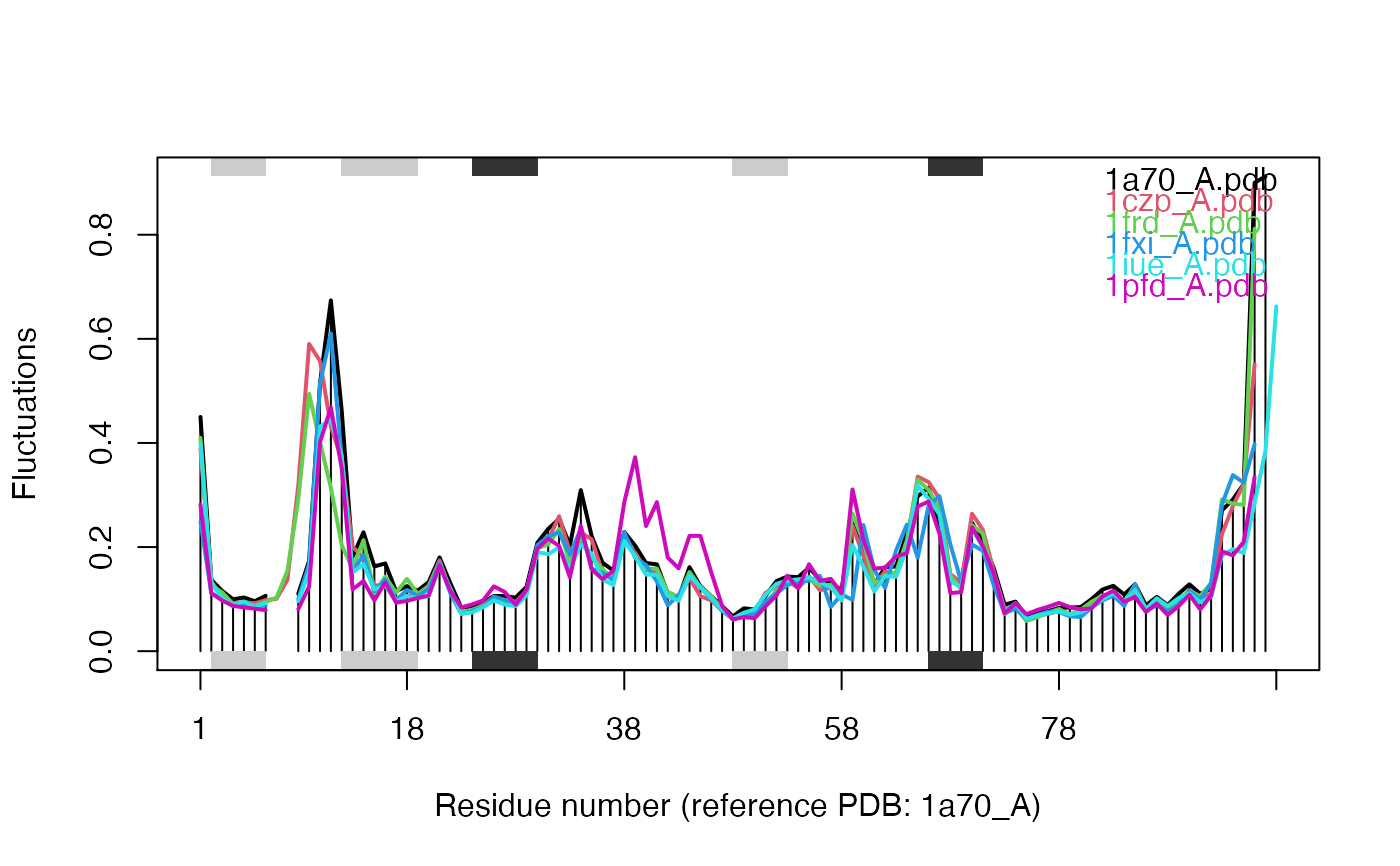
##- Singe structure NMA ## Fetch stucture pdb <- read.pdb( system.file("examples/1hel.pdb", package="bio3d") ) ## Calculate normal modes modes <- nma(pdb)#> Building Hessian... Done in 0.011 seconds. #> Diagonalizing Hessian... Done in 0.078 seconds.#> #> Call: #> nma.pdb(pdb = pdb) #> #> Class: #> VibrationalModes (nma) #> #> Number of modes: #> 387 (6 trivial) #> #> Frequencies: #> Mode 7: 0.018 #> Mode 8: 0.019 #> Mode 9: 0.024 #> Mode 10: 0.025 #> Mode 11: 0.028 #> Mode 12: 0.029 #> #> + attr: modes, frequencies, force.constants, fluctuations, #> U, L, xyz, mass, temp, triv.modes, natoms, call## Visualize modes #m7 <- mktrj.nma(modes, mode=7, file="mode_7.pdb") # \donttest{ ## Needs MUSCLE installed - testing excluded ##- Ensemble NMA if(check.utility("muscle")) { ## Fetch PDB files and split to chain A only PDB files ids <- c("1a70_A", "1czp_A", "1frd_A", "1fxi_A", "1iue_A", "1pfd_A") files <- get.pdb(ids, split = TRUE, path = tempdir()) ## Sequence Alignement pdbs <- pdbaln(files, outfile = tempfile()) ## Normal mode analysis on aligned data modes <- nma(pdbs, rm.gaps=FALSE) ## Plot fluctuation data plot(modes, pdbs=pdbs) }#> Warning: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/1a70.pdb exists. Skipping download#> Warning: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/1czp.pdb exists. Skipping download#> Warning: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/1frd.pdb exists. Skipping download#> Warning: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/1fxi.pdb exists. Skipping download#> Warning: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/1iue.pdb exists. Skipping download#> Warning: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/1pfd.pdb exists. Skipping download#> | | | 0% | |============ | 17% | |======================= | 33% | |=================================== | 50% | |=============================================== | 67% | |========================================================== | 83% | |======================================================================| 100% #> Reading PDB files: #> /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1a70_A.pdb #> /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1czp_A.pdb #> /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1frd_A.pdb #> /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1fxi_A.pdb #> /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1iue_A.pdb #> /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1pfd_A.pdb #> . PDB has ALT records, taking A only, rm.alt=TRUE #> ..... #> #> Extracting sequences #> #> pdb/seq: 1 name: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1a70_A.pdb #> pdb/seq: 2 name: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1czp_A.pdb #> PDB has ALT records, taking A only, rm.alt=TRUE #> pdb/seq: 3 name: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1frd_A.pdb #> pdb/seq: 4 name: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1fxi_A.pdb #> pdb/seq: 5 name: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1iue_A.pdb #> pdb/seq: 6 name: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp9oBdbc/split_chain/1pfd_A.pdb #> #> Details of Scheduled Calculation: #> ... 6 input structures #> ... storing 282 eigenvectors for each structure #> ... dimension of x$U.subspace: ( 300x282x6 ) #> ... coordinate superposition prior to NM calculation #> ... estimated memory usage of final 'eNMA' object: 3.9 Mb #> #> | | | 0% | |============ | 17% | |======================= | 33% | |=================================== | 50% | |=============================================== | 67% | |========================================================== | 83% | |======================================================================| 100%#># }