User guide
This page presents a hands on introduction to Bio3D and the R environment. It is intended to familiarize the user with a few essentials important for getting off to a quick start.
In the examples below we assume that you have successfully installed R and the Bio3D package, by following the installation instructions on the Download page. Extended installation instructions are also available as a PDF for download.
This page contains the following sections:
- Introduction
- Do I need to know R?
- Loading Bio3D
- Finding Help
- Quitting and Bailing out
- Saving your work
- Running the Bio3D Demos
- Extended Tutorals (a.k.a: The Bio3D vignettes)
A number of worked examples are available as extensive Tutorials. You can also view the online Documentation or download the package manual (in PDF format).
Introduction
R is a software environment for statistical computing and graphics. Among other things it includes:
- an effective data handling and storage facility,
- a suite of operators for calculations on arrays, in particular matrices,
- a large, coherent, integrated collection of intermediate tools for data analysis,
- graphical facilities for data analysis and display either on-screen or on hardcopy, and
- a well-developed, simple and effective programming language.
Bio3D is an R package containing utilities to process, organize and explore structure and sequence data. Features include the ability to read and write structure, sequence and dynamic trajectory data, perform atom selection, re-orientation, superposition, rigid core identification, clustering, distance matrix analysis, conservation analysis, principal component analysis (PCA), and normal mode analysis (NMA). Bio3D takes advantage of the extensive graphical and statistical capabilities of the R environment and thus represents a useful framework for exploratory analysis of structural data.
Do I need to know R?
To get the most out of Bio3D you should be somewhat familiar with basic R usage. Some newcomers find this a steep learning curve. However, once you have mastered basic operations with vectors and matrices you should feel confident about getting stuck into using the Bio3D package. We believe that what Bio3D in R lacks in apparent initial user-friendliness, it more than makes up for in power. See our why should I use R page for more discussion.
New to R?
There are now numerous on–line resources that can help you get started using R. A number of these can be found from the main R website at http://www.r-project.org. We particularly like the following:
- Try R: a nifty interactive R tutorial in your web browser
- An introduction to R: The offical R manual
- Learn R: Learn by doing in your web browser (requires free registration)
Loading bio3d
Start R (type R at the command prompt or, on Windows, double click on the R icon) and load the Bio3D package by typing library(bio3d) at the R console prompt. 
Use the command lbio3d() to list the functions within the package. 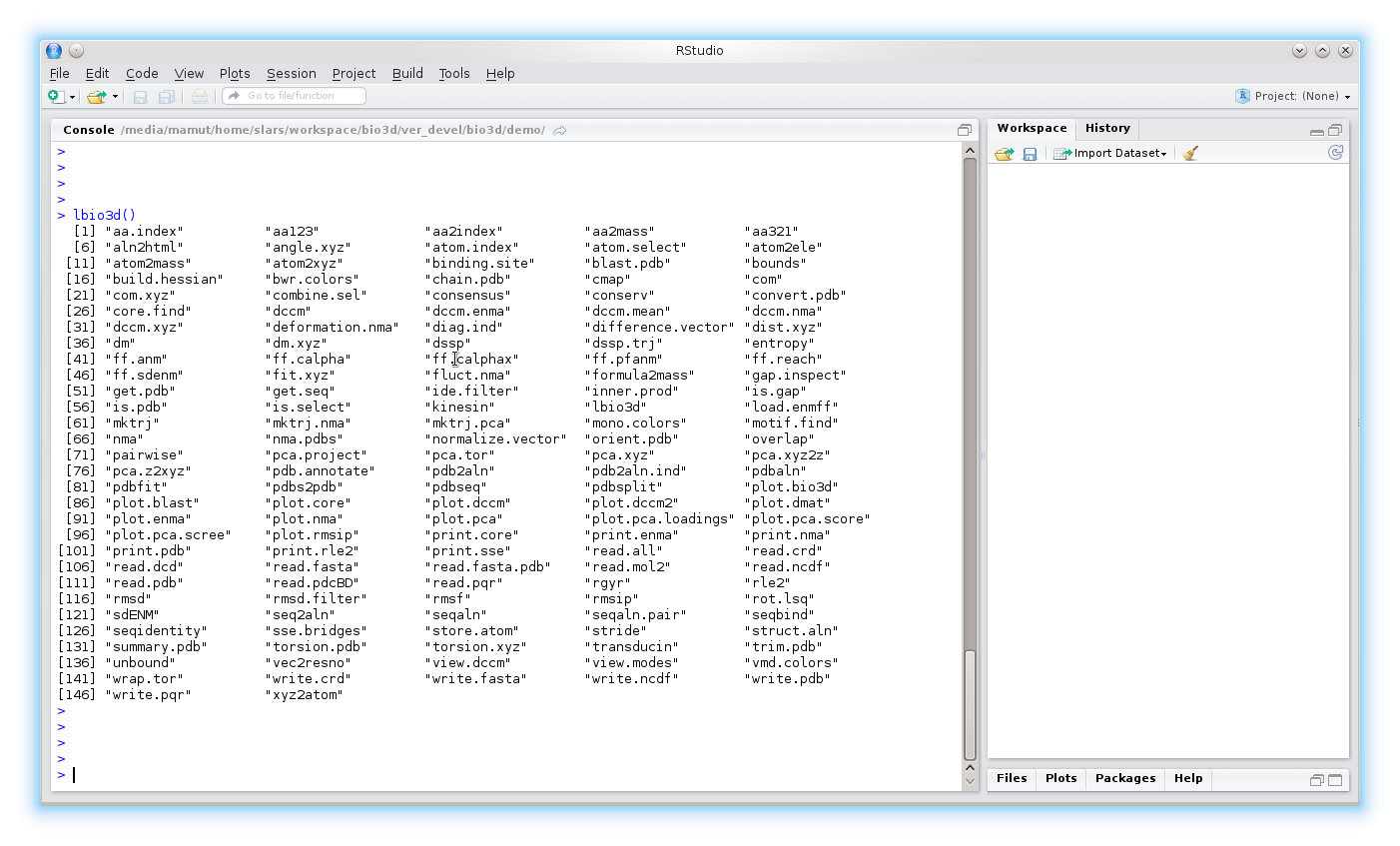
Finding Help
To get help on a particular function try ?function or help(function). For example, ?pca.xyz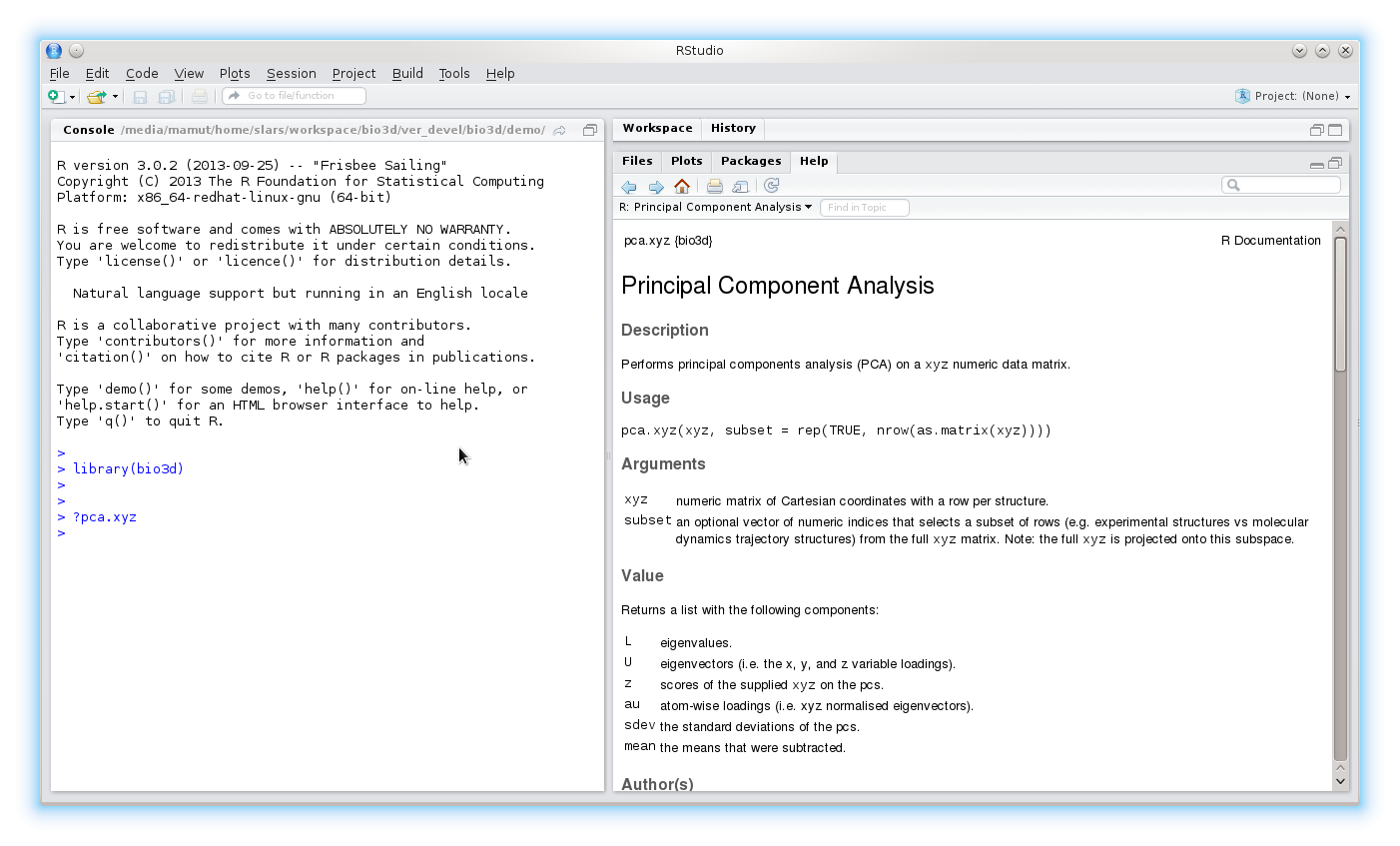
To search the help system for documentation matching a particular word or topic use the command help.search("topic"). For example, help.search("pdb")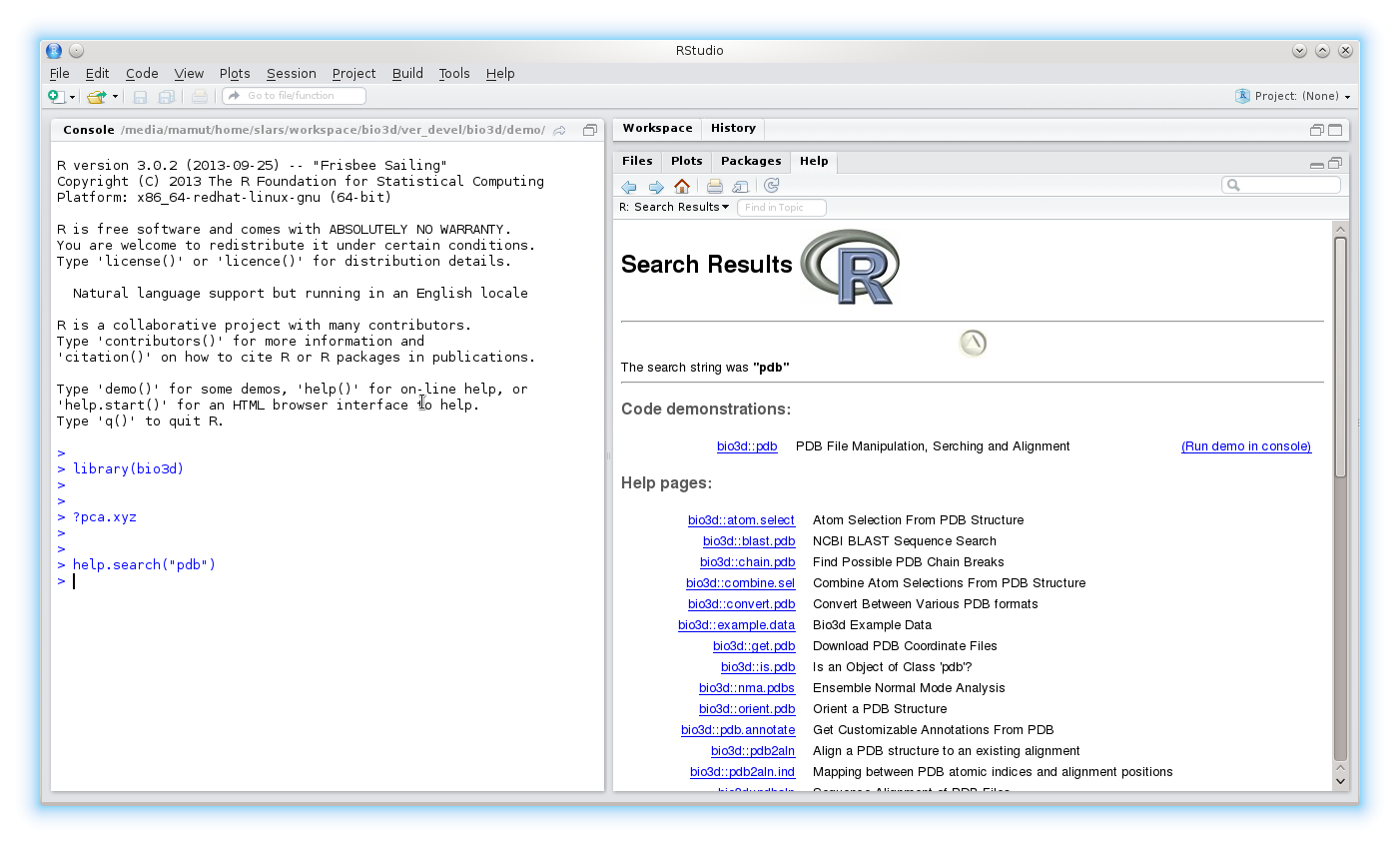
Typing help.start() will start a local HTML interface. After initiating help.start() in a session the ?function commands will open as HTML pages.
To execute examples for a particular function use the command example(function). To run examples for the nma function try example(nma)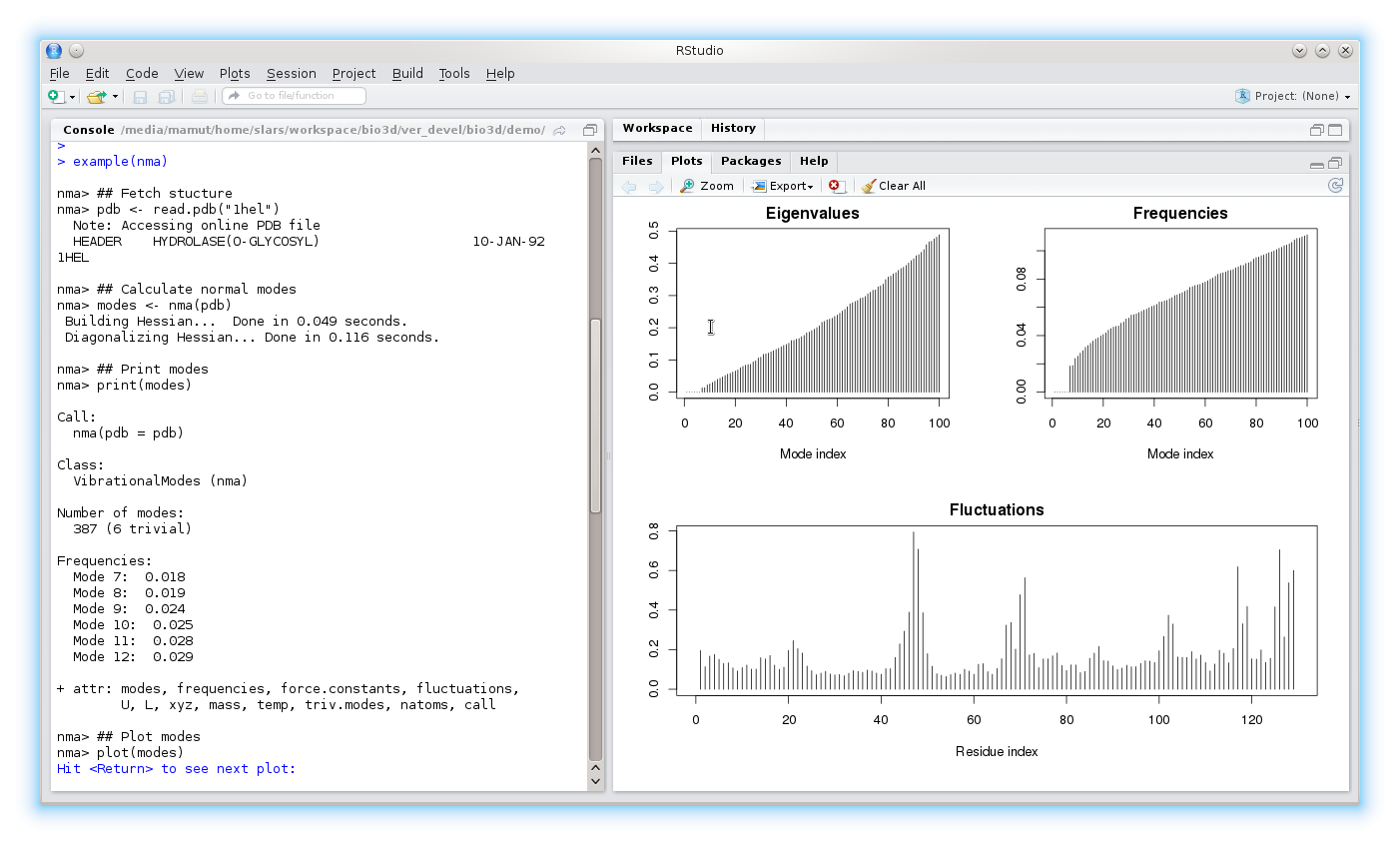
Quitting and Bailing out
R is generally very tolerant, and can be interrupted by Ctrl-C (i.e. hold down the key marked Control and hit the C key). This will interrupt the current operation and return to the R prompt. To quit R all-together, type q()
Note the option to save your work for future use: "save workspace image [y/n/c]".
Answering yes to this question will result in a binary .RData file being written to the current directory. This file contains details of all objects created in your current session. This file is read the next time R is executed in this directory, providing access to all your previous objects. To avoid this, answer "n" at quit time, or erase the .RData file before re-starting R.
Saving your work
You can save an external representation of any R objects to a specified file using the save()command. The objects can be read back from the file at a later time by using the function load(). 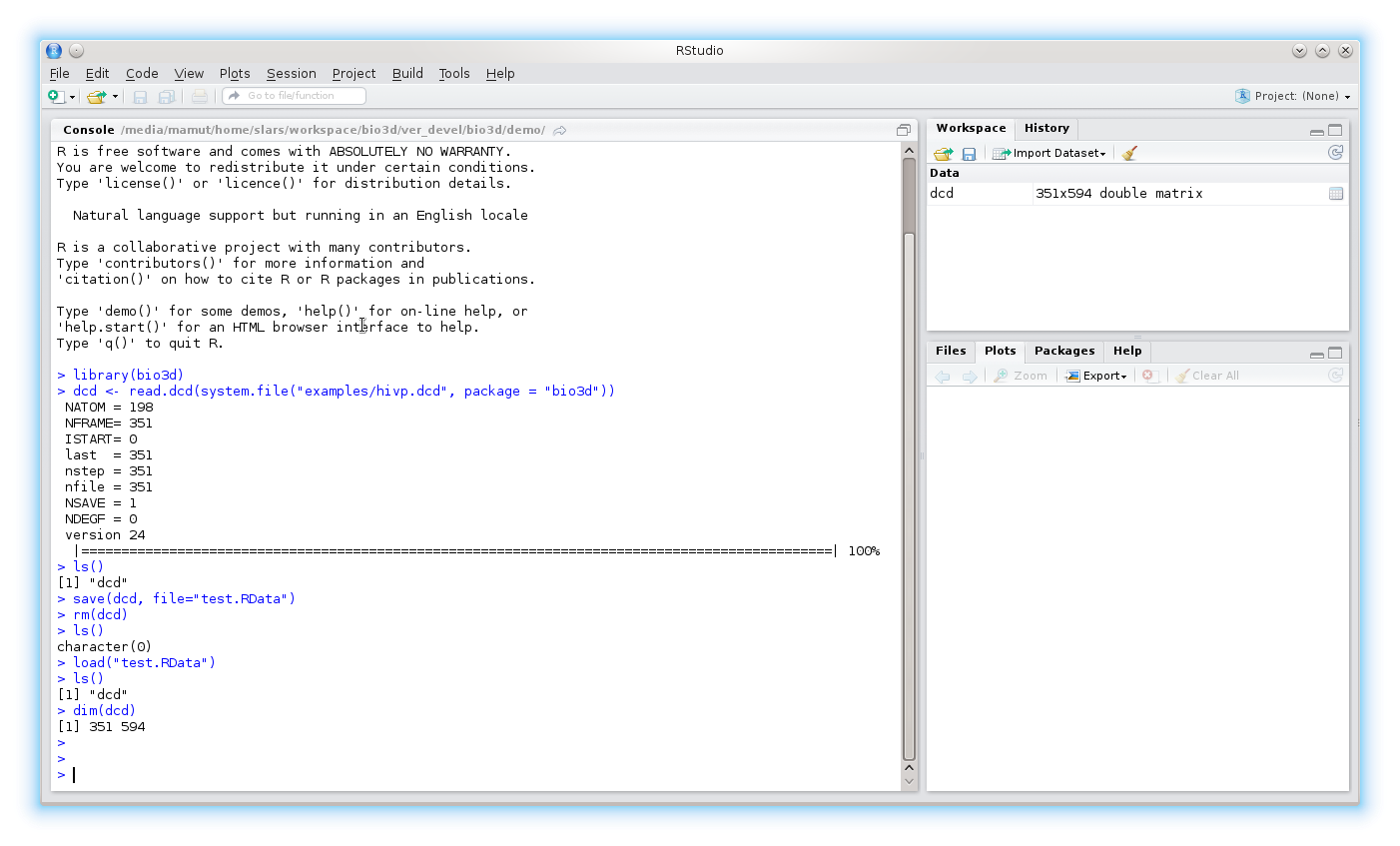
To save all objects in your current session use the save.image() command.
Bio3d Demos
The Bio3D package ships with a number of extended demos that go beyond the example code provided for each function within its documentation.
To see availalbe demos run the command demo(package="bio3d"). Then for example run the command demo("nma"), or demo("md") etc.
Note that these demos will run through extended application of Bio3D functions for a particular task (such as normal mode analysis, or molecular dynamics simulation analysis, etc.). You will be prompted to hit the return key at each step as this will allow you to see the particular functions being called.
Remember that you can get help on any particular function by using the command ?function or help(function) from within R.
Example Function Usage
To better understand how a particular function operates it is often helpful to view and execute an example. Every function within the Bio3D package is documented with example code that you can view by issuing the help command.
Running the command example(function) will directly execute the example for a given function.
In addition, a numer of worked examples are available as so-called package vignettes (basically short tutorials). Currently available vignettes include:
- Installing Bio3D ( PDF | HTML )
- Beginning Structure Analysis with Bio3D ( PDF | HTML )
- Beginning Trajectory Analysis with Bio3D ( PDF | HTML )
- Enhanced Methods for Normal Mode Analysis with Bio3D ( PDF | HTML )
- Comparative sequence and structure analysis with Bio3D ( PDF | HTML )
- Introduction to Correlation Network Analysis with Bio3D ( PDF | HTML )
