Trajectory Analysis with Bio3D
Background
Bio3D1 is an R package that provides interactive tools for the analysis of bimolecular structure, sequence and simulation data. The aim of this document, termed a vignette2 in R parlance, is to provide a brief task-oriented introduction to basic molecular dynamics trajectory analysis with the Bio3D R package (Grant et al. 2006).
Requirements
Detailed instructions for obtaining and installing the Bio3D package on various platforms can be found in the Installing Bio3D vignette available online. This particular vignette was generated using Bio3D version 2.3.0.
Getting Started
Start R, load the Bio3D package and use the command demo("md") to get
a quick feel for some of the tasks that we will be introducing in the
following sections.
library(bio3d)
demo("md")
Side-note:
Note that you will be prompted to hit the RETURN key at each step of
the demo as this will allow you to see the particular functions being
called. Also note that detailed documentation and example code for each
function can be accessed via the help() and example() commands (e.g.
help(read.pdb)). You can also copy and paste any of the example code
from the documentation of a particular function, or indeed this
vignette, directly into your R session to see how things work. You can
also find this documentation
online.
Reading Example Trajectory Data
A number of example data sets are shipped with the Bio3D package. The main purpose of including this data is to allow users to more quickly appreciate the capabilities of various Bio3D functions that would otherwise require potentially time consuming data generation. In the examples below we will input, process and analyze a molecular dynamics trajectory of Human Immunodeficiency Virus aspartic protease (HIVpr). This trajectory is stored in CHARMM/NAMD DCD format and has had all solvent and non C-alpha protein atoms excluded to reduce overall file size.
The code snippet below sets the file paths for the example HIVpr starting structure (pdbfile) and trajectory data (dcdfile).
dcdfile <- system.file("examples/hivp.dcd", package="bio3d")
pdbfile <- system.file("examples/hivp.pdb", package="bio3d")
Side-note:
Note that in the above example the system.file() command returns a
character string corresponding to the file name of a PDB structure
included with the Bio3D package. This is required as users may install
the package in different locations. When using your own input files the
system.file() command will not be required, for example
mydcdfile <- "/path/to/my/data/myfile.dcd"
dcd <- read.dcd(dcdfile)
pdb <- read.pdb(pdbfile)
The read.dcd() and read.pdb() commands processes the input files and
returns their output to the new objects dcd and pdb. We can check
the basic structure of these objects with the following commands:
print(pdb)
##
## Call: read.pdb(file = pdbfile)
##
## Total Models#: 1
## Total Atoms#: 198, XYZs#: 594 Chains#: 2 (values: A B)
##
## Protein Atoms#: 198 (residues/Calpha atoms#: 198)
## Nucleic acid Atoms#: 0 (residues/phosphate atoms#: 0)
##
## Non-protein/nucleic Atoms#: 0 (residues: 0)
## Non-protein/nucleic resid values: [ none ]
##
## Protein sequence:
## PQITLWQRPLVTIKIGGQLKEALLDTGADDTVLEEMSLPGRWKPKMIGGIGGFIKVRQYD
## QILIEICGHKAIGTVLVGPTPVNIIGRNLLTQIGCTLNFPQITLWQRPLVTIKIGGQLKE
## ALLDTGADDTVLEEMSLPGRWKPKMIGGIGGFIKVRQYDQILIEICGHKAIGTVLVGPTP
## VNIIGRNLLTQIGCTLNF
##
## + attr: atom, xyz, helix, sheet, calpha,
## call
print(pdb$xyz)
##
## Total Frames#: 1
## Total XYZs#: 594, (Atoms#: 198)
##
## [1] 51.842 59.784 -6.815 <...> 55.089 55.232 -6.899 [594]
##
## + attr: Matrix DIM = 1 x 594
print(dcd)
##
## Total Frames#: 117
## Total XYZs#: 594, (Atoms#: 198)
##
## [1] 51.842 59.784 -6.815 <...> 27.995 70.42 -33.908 [69498]
##
## + attr: Matrix DIM = 117 x 594
The output of the above function is telling us that we have 351
trajectory frames and 594 coordinates. The trajectory information is
stored in a xyz matrix object with one row per frame and columns for
the Cartesian coordinates (x, y and z columns).
Question:
How many atoms are in the trajectory and PDB files?
Question:
How would you extract the amino acid sequence of the HIVpr system in
1-letter and 3-letter forms? HINT: try help.search("PDB sequence") for
a Bio3D function that might help you.
Side-note:
Note that typically one works with trajectory files that contain all
protein atoms, or at the very least all backbone atoms. Solvent however
can often be excluded prior to Bio3D input - it just depends upon your
particular analysis questions. For example, we are not able to analyze
Hydrogen bonding patterns or details of water occupancy with the
currently inputed data.
Trajectory Frame Superposition
In this simple example we select all C-alpha atoms for trajectory frame superposition.
ca.inds <- atom.select(pdb, elety="CA")
The returned ca.inds object is a list containing atom and xyz numeric
indices that we can now use to superpose all frames of the trajectory on
the selected indices (in this case corresponding to all alpha Carbon
atoms). For this we will with the fit.xyz() function.
xyz <- fit.xyz(fixed=pdb$xyz, mobile=dcd,
fixed.inds=ca.inds$xyz,
mobile.inds=ca.inds$xyz)
The above command performs the actual superposition and stores the new
coordinates in the matrix object xyz. Note that the dimensions (i.e.
number of rows and columns, which correspond to frames and coordinates
respectively) of xyz match those of the input trajectory:
dim(xyz) == dim(dcd)
## [1] TRUE TRUE
Question:
How would you fit trajectory frames on the Calpha atoms of residues 24
to 27 and 85 to 90 in both chains? HINT: See the example section of
help(atom.select).
Question:
Would you expect the alternate fitting suggested above to alter your later results? HINT: You can come back to this question later after going through the other sections.
Side-note:
A simple way to obtain the average structure from your fitted trajectory
is to use the following command apply(xyz,2,mean).
Root Mean Square Deviation (RMSD)
RMSD is a standard measure of structural distance between coordinate
sets and is implemented in the Bio3D function rmsd().
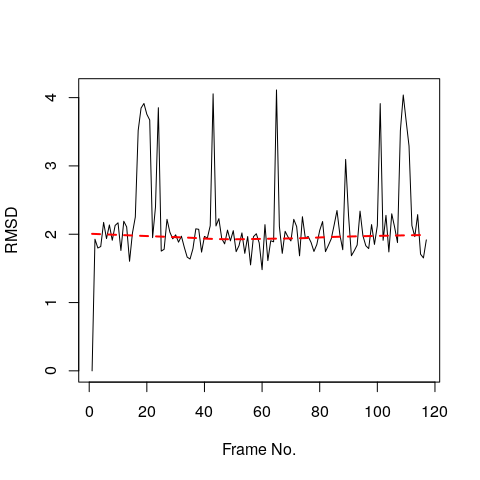
rd <- rmsd(xyz[1,ca.inds$xyz], xyz[,ca.inds$xyz])
plot(rd, typ="l", ylab="RMSD", xlab="Frame No.")
points(lowess(rd), typ="l", col="red", lty=2, lwd=2)
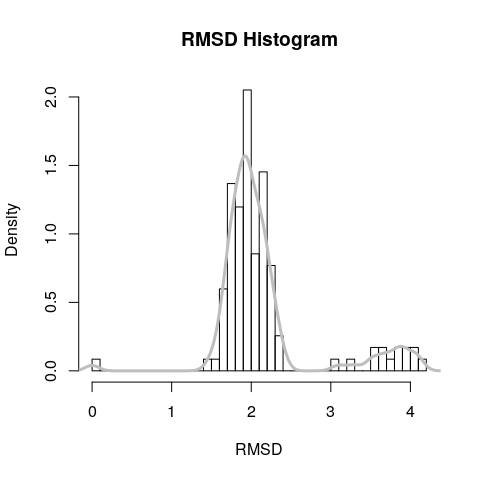
A quick histogram can be useful for examining the distribution of RMSD values.
hist(rd, breaks=40, freq=FALSE, main="RMSD Histogram", xlab="RMSD")
lines(density(rd), col="gray", lwd=3)
summary(rd)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.000 1.834 1.967 2.151 2.167 4.112
Question:
How would you calculate the pairwise RMSD between all frames?
Question:
What would be a good way to visualize and further analyze such a
pairwise RMSD matrix? HINT: Have a look at the NMA vignette for some,
hopefully, inspiring plots?
Root Mean Squared Fluctuations (RMSF)
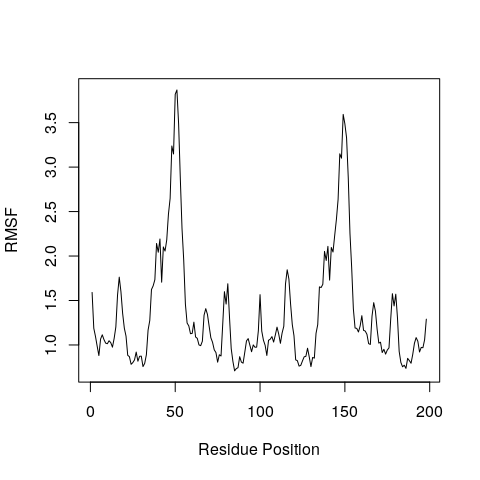
RMSF is an often used measure of conformational variance and is
implemented in the Bio3D function rmsf(). This analysis will highlight
the portions of structure that are fluctuating from their mean structure
the most (and least).
rf <- rmsf(xyz[,ca.inds$xyz])
plot(rf, ylab="RMSF", xlab="Residue Position", typ="l")
Question:
If you had multiple simulations (which by the way we greatly encourage),
how would you plot these on the same graph? HINT: See help(points).
Question:
If you noticed differences at particular sites (e.g. in the presence or
absence of a ligand) how would you go about addressing the significance
of these differences? HINT: this is why we always encourage a multiple
simulation approach and implement Bio3D in one of the most advanced
statistical analysis packages available.
Principal Component Analysis
PCA can be employed to examine the relationship between different
conformations sampled during the trajectory and is implemented in the
Bio3D functions pca.xyz() and pca.tor(). The application of PCA to
both distributions of experimental structures and molecular dynamics
trajectories will be covered in detail in other vignettes. Briefly, we
will note here that this method can provide considerable insight into
the nature of conformational differences with the resulting principal
components (orthogonal eigenvectors) describing the axes of maximal
variance of the distribution of structures. Projection of the
distribution onto the subspace defined by the largest principal
components results in a lower dimensional representation of the
structural dataset (see Figure 4). The percentage of the total mean
square displacement (or variance) of atom positional fluctuations
captured in each dimension is characterized by their corresponding
eigenvalue (see Figure 4D). Experience suggests that 3–5 dimensions
are often sufficient to capture over 70 percent of the total variance in
a given family of experimental structures or indeed a standard molecular
dynamics trajectory. Thus, a handful of principal components are
sufficient to provide a useful description while still retaining most of
the variance in the original distribution (Grant et al. 2006).
A quick overview of the results of pca.xyz() can be obtained by
calling plot.pca()
pc <- pca.xyz(xyz[,ca.inds$xyz])
plot(pc, col=bwr.colors(nrow(xyz)) )
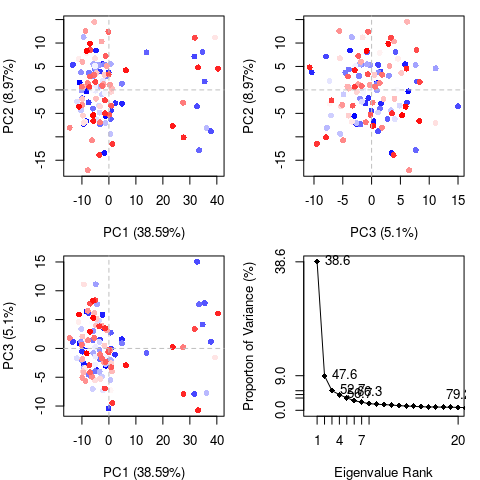
Note that there are distinct groupings of conformations along the PC1 plane (one centered around -30 and a second, larger grouping, at +5). The continuous color scale (from blue to whit to red) indicates that there are periodic jumps between these conformers throughout the trajectory. Below we perform a quick clustering in PC-space to further highlight these distinct conformers.
hc <- hclust(dist(pc$z[,1:2]))
grps <- cutree(hc, k=2)
plot(pc, col=grps)
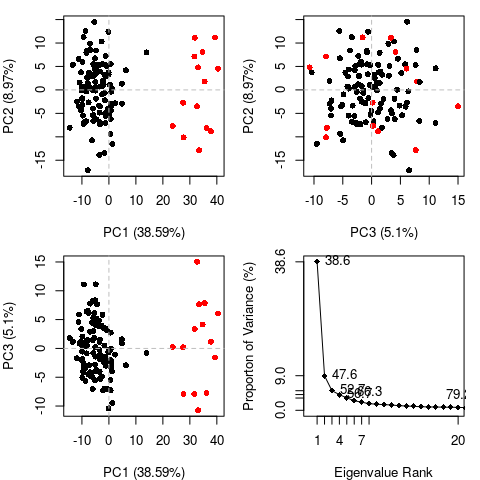
Question
How can we find out which frames correspond to the distinct groups along
PC1? HINT: Which variable/object created above tells us about cluster
membership?
Bellow we call plot.bio3d() to examine the contribution of each
residue to the first two principal components.
plot.bio3d(pc$au[,1], ylab="PC1 (A)", xlab="Residue Position", typ="l")
points(pc$au[,2], typ="l", col="blue")
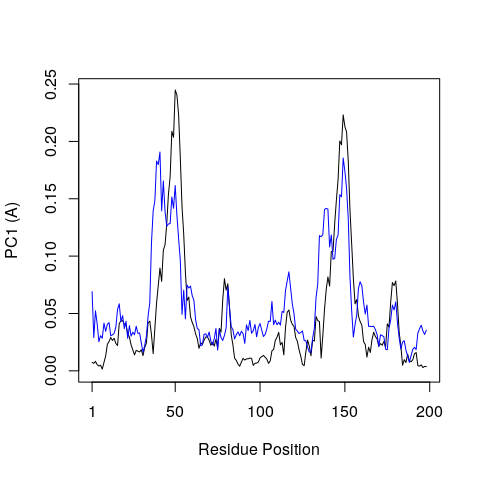
Question:
Why do you think there might be two major peaks in our RMSF plot?
To further aid interpretation, a PDB format trajectory can be produced that interpolates between the most dissimilar structures in the distribution along a given principal component. This involves dividing the difference between the conformers into a number of evenly spaced steps along the principal components, forming the frames of the output multi-model PDB trajectory. Such trajectories can be directly visualized in a molecular graphics program, such as VMD (Humphrey 1996). Furthermore, the interpolated structures can be analyzed for possible domain and shear movements with other Bio3D functions, or used as initial seed structures for reaction path refinement methods (note you will likely want to perform all heavy atom PCA for such applications).
p1 <- mktrj.pca(pc, pc=1, b=pc$au[,1], file="pc1.pdb")
p2 <- mktrj.pca(pc, pc=2,b=pc$au[,2], file="pc2.pdb")
You can also write these trajectory’s as AMBER NetCDF format files with
the write.ncdf function. To view the PDB trajectories in VMD just open
the files in the normal way and display as tube representation for
example (see figure below).
write.ncdf(p1, "trj_pc1.nc")
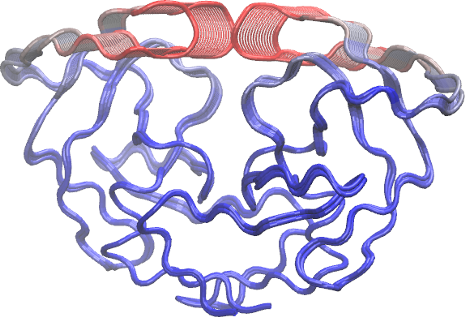
Question:
Which regions are the most dynamic and why?
Cross-Correlation Analysis
The extent to which the atomic fluctuations/displacements of a system
are correlated with one another can be assessed by examining the
magnitude of all pairwise cross-correlation coefficients. The Bio3D
dccm() function returns a matrix of all atom-wise cross-correlations
whose elements may be displayed in a graphical representation frequently
termed a dynamical cross-correlation map, or DCCM.
cij<-dccm(xyz[,ca.inds$xyz])
plot(cij)
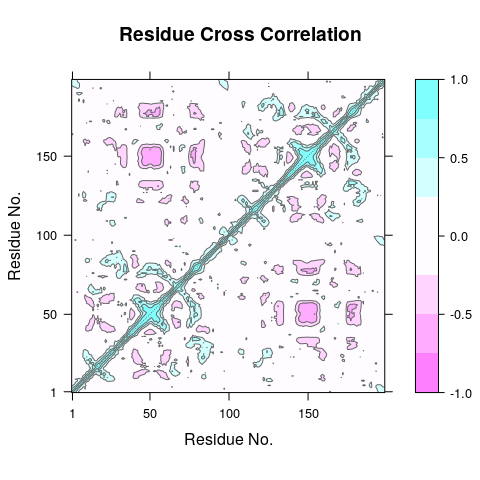
Question:
What do the off-diagonal regions of negative correlation correspond to
in this plot and which regions are involved? HINT: Negative values
typical indicate regions that move in opposite directions.
A 3D visualization of these correlations can be provided through the
function pymol.dccm()
# View the correlations in pymol
pymol.dccm(cij, pdb, type="launch")
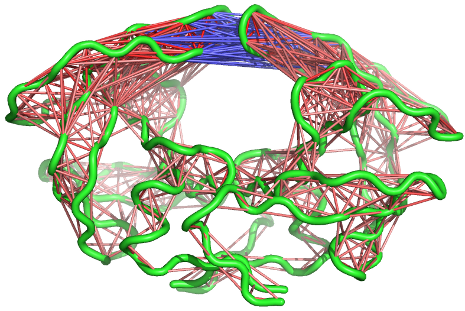
See also the Enhanced Methods for Normal Mode Analysis for additional
visualization examples. Also you might want to checkout the
Comparative Analysis of Protein Structures vignette for relating
results like these to available experimental data. The logical expansion
of this analysis is described in the Correlation Network Analysis
vignette.
Where to Next
If you have read this far, congratulations! We are ready to have some
fun and move to other package
vignettes that describe more
interesting analysis including Correlation Network Analysis (where we
will build and dissect dynamic networks form different correlated motion
data), enhanced methods for Normal Mode Analysis (where we will explore
the dynamics of large protein families and superfamilies), and advanced
Comparative Structure Analysis (where we will mine available
experimental data and supplement it with simulation results to map the
conformational dynamics and coupled motions of proteins).
Document Details
This document is shipped with the Bio3D package in both R and PDF formats. All code can be extracted and automatically executed to generate Figures and/or the PDF with the following commands:
library(rmarkdown)
render("Bio3D_md.Rmd", "all")
Information About the Current Bio3D Session
sessionInfo()
## R version 3.3.1 (2016-06-21)
## Platform: x86_64-redhat-linux-gnu (64-bit)
## Running under: Fedora 24 (Twenty Four)
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] rmarkdown_1.0 bio3d_2.3-0
##
## loaded via a namespace (and not attached):
## [1] magrittr_1.5 formatR_1.4 htmltools_0.3.5 tools_3.3.1
## [5] parallel_3.3.1 yaml_2.1.13 Rcpp_0.12.7 codetools_0.2-14
## [9] stringi_1.1.1 grid_3.3.1 knitr_1.14 stringr_1.0.0
## [13] digest_0.6.10 lattice_0.20-33 evaluate_0.9
References
Grant, B.J., A.P.D.C Rodrigues, K.M. Elsawy, A.J. Mccammon, and L.S.D. Caves. 2006. “Bio3d: An R Package for the Comparative Analysis of Protein Structures.” Bioinformatics 22: 2695–6.
Humphrey, et al., W. 1996. “VMD: Visual Molecular Dynamics.” J. Mol. Graph 14: 33–38.
-
The latest version of the package, full documentation and further vignettes (including detailed installation instructions) can be obtained from the main Bio3D website: http://thegrantlab.org/bio3d/ ↩
-
This vignette contains executable examples, see
help(vignette)for further details. ↩
