Computational & Theoretical Studies of Cellular Hardware
29/09/14 14:53
At the interface of biochemistry, biophysics and bioinformatics the unique insight gained from our research is tightly coupled to a wide range of biochemical and biophysical experiments. Our ultimate goal is to exploit these collaborative efforts to facilitate the fields of protein design and drug discovery.
Read More...
Molecular Motors & Molecular Switches
29/09/14 14:53
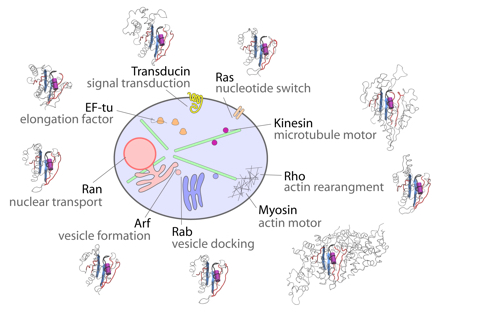
Molecular motors and switches comprise a set of related nucleotide binding proteins that lie at the heart of life's processes, from the division and growth of cells to the muscular movement of organisms. They represent attractive targets for drug design as their aberrant function is associated with many diseases including cancer.
Our research aims to decipher the physical mechanisms by which these fascinating natural nanomachines function and how their dysfunction is related to disease. Our recent findings on these systems include how to rationally engineer faster velocity molecular motors and inhibit molecular switches that are malfunctioning in more than 20% of all human cancers.
Our research aims to decipher the physical mechanisms by which these fascinating natural nanomachines function and how their dysfunction is related to disease. Our recent findings on these systems include how to rationally engineer faster velocity molecular motors and inhibit molecular switches that are malfunctioning in more than 20% of all human cancers.
Molecular Motors & the Cytoskeleton
29/09/14 14:53
We are fascinated by molecular motion, particularly that of nature’s motor proteins: kinesin, myosin and dynein. Together with their cellular highways, microtubules and actin filaments, these proteins drive the beating of sperm, the division of cells and the muscular movement of organisms. Our research is focused on deciphering how these mighty molecular motors work, and how to manipulate them for industrial and medical advantage.
A principal aim in the study of molecular motors, is a detailed understanding of how chemical energy, derived from the hydrolysis of ATP, is converted into mechanical energy. Read More...
Conformational Selection & Allostery in Molecular Switches
29/09/14 14:53
Our atomistic molecular simulations indicate that these enzymes harbor an intrinsic susceptibility to sample multiple conformational states regardless of the bound nucleotide. These results indicate that conformational changes in these enzymes may be best described by a population-shift mechanism rather than by the popular ligand induced-fit on/off switch model. Read More...
Methods for Computer Aided Drug Design
29/09/14 14:53
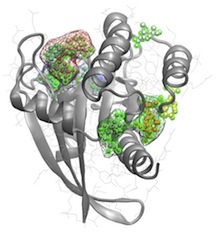
We are developing a more sophisticated methodology that exploits flexible target structures, allosteric pocket identification and evolutionary analysis. Merging these new methods represents an important advance that will greatly facilitate the design of selective inhibitors.
Read More...
Integrating Protein Structural Dynamics and Evolutionary Analysis
29/09/14 14:53
The Grant lab are lead developers of Bio3D, an open source R package for the exploratory analysis of protein sequence, structure and dynamic trajectory data.
Features of the package include extensive graphical and statistical capabilities, atom selection, rigid core superposition, clustering, distance matrix analysis, structure and sequence conservation analysis and principal component analysis.
Read More...
Features of the package include extensive graphical and statistical capabilities, atom selection, rigid core superposition, clustering, distance matrix analysis, structure and sequence conservation analysis and principal component analysis.
Read More...
Target Selection in Structural Genomics
29/09/14 14:53
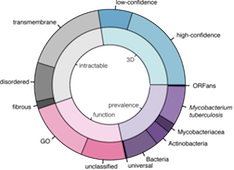
The system enables structural biologists to select targets from their genomic sequences of interest, according to their research needs.
Read More...
Channel Gating in Nicotinic Acetylcholine Receptors
29/09/14 14:52
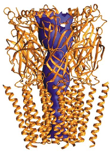
Molecular simulation methods are currently being used to probe how conformational changes might propagate from the ligand-binding site to the pore domain and result in channel gating. Read More...
Recent Publications
29/09/14 14:52
Mapping the processivity determinants of the kinesin-3 motor domain.
Scarabelli, Soppina, Yao, Atherton, Moores, Verhey, Grant (2015)
Biophys J. 109, 1537–1540.
( Abstract | PubMed | PDF )
Enrichment of druggable conformations from apo protein structures using cosolvent-accelerated molecular dynamics.
Kalenkiewicz, Grant, Yang (2015)
Biology 4, 344-366
( Abstract | PubMed | PDF )
Integrating protein structural dynamics and evolutionary analysis with Bio3D.
Skjærven, Yao, Scarabelli, Grant (2014)
BMC Bioinformatics 15, 399
( Abstract | PubMed | PDF )
Kinesin-5 allosteric inhibitors uncouple the dynamics of nucleotide, microtubule and neck-linker binding sites.
Scarabelli, Grant (2014)
Biophys J 107, 2204–2213
( Abstract | PubMed | PDF )
Mapping the structural and dynamical features of kinesin motor domains.
Scarabelli, Grant (2013)
PLoS Comput Biol 9, e1003329
( Abstract | PubMed | PDF )
Domain opening and dynamic coupling in the alpha subunit of heterotrimeric G proteins.
Yao, Grant (2013)
Biophys J 105, L08–L10
( Abstract | PubMed | PDF )
Identification of potential small molecule binding pockets on Rho family GTPases.
Ortiz Sanchez, Nichols, Sayyah, Heller Brown, McCammon, Grant (2012)
PLoS One 7, e40809
( Abstract | PubMed | PDF )
Calcium binding and allosteric signaling mechanisms for Sarcoplasmic reticulum Ca2+ ATPase.
Kekenes-Huskey, Metzger, Grant, McCammon (2012)
Protein Science 10, 1429-43
( Abstract | PubMed | PDF )
Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire.
Ostrov, Grant, Pompeu, Sidney, Harndahl, Southwood, Oseroff, Lu, Jakoncic, de Oliveira, Yang, Mei, Shi, Shabanowitz, English, Wriston, Lucas, Phillips, Mallal, Grey, Sette, Hunt, Buus, Peters (2012)
PNAS 109, 9959-64.
( Abstract | PubMed | PDF )
LeuT conformational sampling utilizing accelerated molecular dynamics and principal component analysis.
Thomas, Gedeon, Grant, Madura (2012)
Biophys J 103, L1-3
( Abstract | PubMed | PDF )
Electrostatically biased binding of kinesin to microtubules.
Grant*, Gheorghe*, Zheng, Alonso, Huber, Dlugosz, McCammon, Cross (2011)
PLoS Biology 9, e1001207
( Abstract | PubMed | PDF | Press: 1, 2, 3)
Induced-fit or conformational selection? The role of the semi-closed state in the Maltose Binding Protein.
Bucher, Grant, McCammon (2011)
Biochemistry 50, 10530–10539
( Abstract | PubMed | PDF )
Novel allosteric sites on Ras for lead generation.
Grant*, Lukman*, Hocker, Sayyah, Heller Brown, McCammon, Gorfe (2011)
PLoS One 6, e25711
( Abstract | PubMed | PDF )
Large-scale conformational changes of Trypanosoma cruzi proline racemase predicted by accelerated molecular dynamics simulation.
Oliveira*, Grant*, Zhou, McCammon (2011)
PLoS Comput Biol 7, e1002178.
( Abstract | PubMed | PDF )
Accessing a hidden conformation of the maltose binding protein with accelerated molecular dynamics.
Bucher, Grant, Markwick, McCammon (2011)
PLoS Comput Biol 7, e1002004
( Abstract | PubMed | PDF )
Conformational sampling and nucleotide-dependent transitions of the GroEL subunit probed by unbiased molecular dynamics simulations.
Skjaerven, Grant, Muga, Teigen, McCammon, Reuter and Martinez (2011)
PLoS Comput Biol 7, e1002004
( Abstract | PubMed | PDF )
Conformational selection in G-proteins: lessons from Ras and Rho.
Grant, Mccammon, Gorfe (2010)
Biophys J 99, L87-L89
( Abstract | PubMed | PDF )
The distinct conformational dynamics of K-Ras and H-Ras A59G.
Lukman*, Grant*, Gorfe, Mccammon (2010)
PLoS Comput Biol 6, e1000922
( Abstract | PubMed | PDF )
Large conformational changes in proteins: signaling and other functions.
Grant, Gorfe, Mccammon (2010)
Current Opinion in Structural Biology 20, 142-147
( Abstract | PubMed | PDF )
A transition path ensemble study reveals a linchpin role for Mg2+ during rate-limiting ADP release from protein kinase A.
Khavrutskii, Grant, Taylor, Mccammon (2009)
Biochemistry 48, 11532-11545
( Abstract | PubMed | PDF )
Ras conformational switching: simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics.
Grant*, Gorfe*, Mccammon (2009)
PLoS Comput Biol 5, e1000325
( Abstract | PubMed | PDF )
Read More...
Scarabelli, Soppina, Yao, Atherton, Moores, Verhey, Grant (2015)
Biophys J. 109, 1537–1540.
( Abstract | PubMed | PDF )
Enrichment of druggable conformations from apo protein structures using cosolvent-accelerated molecular dynamics.
Kalenkiewicz, Grant, Yang (2015)
Biology 4, 344-366
( Abstract | PubMed | PDF )
Integrating protein structural dynamics and evolutionary analysis with Bio3D.
Skjærven, Yao, Scarabelli, Grant (2014)
BMC Bioinformatics 15, 399
( Abstract | PubMed | PDF )
Kinesin-5 allosteric inhibitors uncouple the dynamics of nucleotide, microtubule and neck-linker binding sites.
Scarabelli, Grant (2014)
Biophys J 107, 2204–2213
( Abstract | PubMed | PDF )
Mapping the structural and dynamical features of kinesin motor domains.
Scarabelli, Grant (2013)
PLoS Comput Biol 9, e1003329
( Abstract | PubMed | PDF )
Domain opening and dynamic coupling in the alpha subunit of heterotrimeric G proteins.
Yao, Grant (2013)
Biophys J 105, L08–L10
( Abstract | PubMed | PDF )
Identification of potential small molecule binding pockets on Rho family GTPases.
Ortiz Sanchez, Nichols, Sayyah, Heller Brown, McCammon, Grant (2012)
PLoS One 7, e40809
( Abstract | PubMed | PDF )
Calcium binding and allosteric signaling mechanisms for Sarcoplasmic reticulum Ca2+ ATPase.
Kekenes-Huskey, Metzger, Grant, McCammon (2012)
Protein Science 10, 1429-43
( Abstract | PubMed | PDF )
Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire.
Ostrov, Grant, Pompeu, Sidney, Harndahl, Southwood, Oseroff, Lu, Jakoncic, de Oliveira, Yang, Mei, Shi, Shabanowitz, English, Wriston, Lucas, Phillips, Mallal, Grey, Sette, Hunt, Buus, Peters (2012)
PNAS 109, 9959-64.
( Abstract | PubMed | PDF )
LeuT conformational sampling utilizing accelerated molecular dynamics and principal component analysis.
Thomas, Gedeon, Grant, Madura (2012)
Biophys J 103, L1-3
( Abstract | PubMed | PDF )
Electrostatically biased binding of kinesin to microtubules.
Grant*, Gheorghe*, Zheng, Alonso, Huber, Dlugosz, McCammon, Cross (2011)
PLoS Biology 9, e1001207
( Abstract | PubMed | PDF | Press: 1, 2, 3)
Induced-fit or conformational selection? The role of the semi-closed state in the Maltose Binding Protein.
Bucher, Grant, McCammon (2011)
Biochemistry 50, 10530–10539
( Abstract | PubMed | PDF )
Novel allosteric sites on Ras for lead generation.
Grant*, Lukman*, Hocker, Sayyah, Heller Brown, McCammon, Gorfe (2011)
PLoS One 6, e25711
( Abstract | PubMed | PDF )
Large-scale conformational changes of Trypanosoma cruzi proline racemase predicted by accelerated molecular dynamics simulation.
Oliveira*, Grant*, Zhou, McCammon (2011)
PLoS Comput Biol 7, e1002178.
( Abstract | PubMed | PDF )
Accessing a hidden conformation of the maltose binding protein with accelerated molecular dynamics.
Bucher, Grant, Markwick, McCammon (2011)
PLoS Comput Biol 7, e1002004
( Abstract | PubMed | PDF )
Conformational sampling and nucleotide-dependent transitions of the GroEL subunit probed by unbiased molecular dynamics simulations.
Skjaerven, Grant, Muga, Teigen, McCammon, Reuter and Martinez (2011)
PLoS Comput Biol 7, e1002004
( Abstract | PubMed | PDF )
Conformational selection in G-proteins: lessons from Ras and Rho.
Grant, Mccammon, Gorfe (2010)
Biophys J 99, L87-L89
( Abstract | PubMed | PDF )
The distinct conformational dynamics of K-Ras and H-Ras A59G.
Lukman*, Grant*, Gorfe, Mccammon (2010)
PLoS Comput Biol 6, e1000922
( Abstract | PubMed | PDF )
Large conformational changes in proteins: signaling and other functions.
Grant, Gorfe, Mccammon (2010)
Current Opinion in Structural Biology 20, 142-147
( Abstract | PubMed | PDF )
A transition path ensemble study reveals a linchpin role for Mg2+ during rate-limiting ADP release from protein kinase A.
Khavrutskii, Grant, Taylor, Mccammon (2009)
Biochemistry 48, 11532-11545
( Abstract | PubMed | PDF )
Ras conformational switching: simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics.
Grant*, Gorfe*, Mccammon (2009)
PLoS Comput Biol 5, e1000325
( Abstract | PubMed | PDF )
Read More...
