Computational & Theoretical Studies of Cellular Hardware
Modern biology is emerging from an Linnaean phase, where genetic and biochemical approaches have assembled a growing inventory of cellular hardware, to a more mechanistic phase, where new insights will come from detailed studies of the dynamics and interactions of these molecules and their complexes.
Artistic representation of a eukaryotic cell's cytoplasm produced by David S. Goodsell Scripps Institute, San Diego.
Gaining a detailed understanding of the inner workings of cells given the complexity of this hardware inventory of gene products is a seemingly overwhelming endeavor. However it is already clear that the origin of complex life forms by evolution has simplified the task. For example, although the human genome encodes over 900 nucleotide binding proteins, many of these have much in common with each other due to their evolution from a common ancestor.
One such group, called the P-loop NTPases, comprise functionally diverse motors, switches and regulators that together account for more than 5% of the human proteome. Members of this large superfamily share many mechanistic commonalities. Hence we believe that detailed knowledge obtained from intensive computational and experimental studies of representative molecules can provide informative general principles about how whole sets of key proteins work and in turn illuminate how evolution has tailored common underlying features to distinct cellular roles.
Research in the Grant lab focuses on the development and application of state-of-the-art computational and theoretical techniques to investigate the structure, dynamics and interactions of these fascinating motors, switches and regulators. All our projects have on-going collaborations with wet-lab cell biologists, biophysicists and biochemists. Together we are deciphering the molecular mechanisms by which cytoskeletal motors and related G-proteins function and how their dysfunction is related to disease.
Probing functional conformational changes in molecular switches with accelerated molecular dynamics simulation.
Probing functional protein-protein interactions with Brownian dynamics simulation
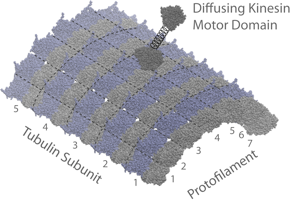
We are also working on extending the theoretical and computational treatment of biomolecules to larger assemblages of proteins including kinesin-microtubule interactions and substrate handling in signaling enzyme complexes. Our ultimate goal is to advance these efforts to facilitate protein design and drug discovery.
<< Back to Research Overview
