Ensemble Normal Mode Analysis with All-Atom ENM
Usage
"aanma"(pdbs, fit = TRUE, full = FALSE, subspace = NULL, rm.gaps = TRUE, ligand = FALSE, outpath = NULL, gc.first = TRUE, ncore = NULL, ...)
Arguments
- pdbs
- an ‘pdbs’ object as obtained from
read.all. - fit
- logical, if TRUE C-alpha coordinate based superposition is performed prior to normal mode calculations.
- full
- logical, if TRUE return the complete, full structure, ‘nma’ objects.
- subspace
- number of eigenvectors to store for further analysis.
- rm.gaps
- logical, if TRUE obtain the hessian matrices for only atoms in the aligned positions (non-gap positions in all aligned structures). Thus, gap positions are removed from output.
- ligand
- logical, if TRUE ligand molecules are also included in the calculation.
- outpath
- character string specifing the output directory to which the PDB structures should be written.
- gc.first
- logical, if TRUE will call gc() first before mode calculation
for each structure. This is to avoid memory overload when
ncore > 1. - ncore
- number of CPU cores used to do the calculation.
- ...
- additional arguments to
aanma.
Value
-
Returns a list of ‘nma’ objects (
- fluctuations
- a numeric matrix containing aligned atomic fluctuations with one row per input structure.
- rmsip
- a numeric matrix of pair wise RMSIP values (only the ten lowest frequency modes are included in the calculation).
- U.subspace
- a three-dimensional array with aligned eigenvectors (corresponding to the subspace defined by the first N non-trivial eigenvectors (‘U’) of the ‘nma’ object).
- L
- numeric matrix containing the raw eigenvalues with one row per input structure.
- full.nma
- a list with a
nmaobject for each input structure (available only whenfull=TRUE).
outmodes is provided
and is not ‘calpha’) or an ‘enma’ object with the following
components:
Description
Perform normal mode analysis (NMA) on an ensemble of aligned protein structures using all-atom elastic network model (aaENM).
Details
This function builds elastic network model (ENM) using all heavy
atoms and performs subsequent normal mode analysis (NMA) on a set of
aligned protein structures obtained with function read.all.
The main purpose is to automate ensemble normal mode analysis using
all-atom ENMs.
By default, the effective Hessian for all C-alpha atoms is calculated
based on the Hessian built from all heavy atoms (including ligand atoms if
ligand=TRUE). Returned values include aligned mode vectors and
(when full=TRUE) a list containing the full ‘nma’ objects
one per each structure. When ‘rm.gaps=TRUE’ the unaligned atoms
are ommited from output. With default arguments ‘rmsip’ provides
RMSIP values for all pairwise structures.
When outmodes is provided and is not ‘calpha’
(e.g. ‘noh’. See aanma for more details), the
function simply returns a list of ‘nma’ objects, one per each
structure, and no aligned mode vector is returned. In this case, the
arguments full, subspace, and rm.gaps are ignored.
This is equivalent to a wrapper function repeatedly calling
aanma.
Examples
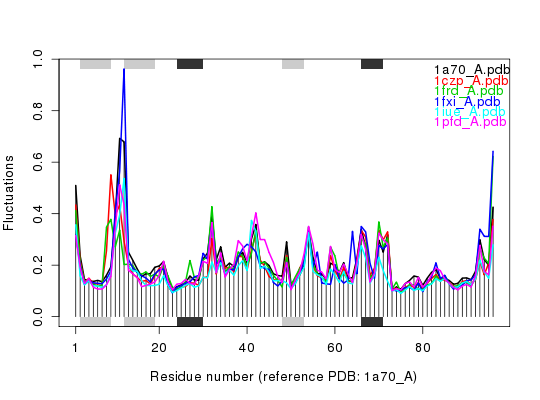
# Needs MUSCLE installed - testing excluded ## Fetch PDB files and split to chain A only PDB files ids <- c("1a70_A", "1czp_A", "1frd_A", "1fxi_A", "1iue_A", "1pfd_A") files <- get.pdb(ids, split = TRUE, path = tempdir())|======================================================================| 100%## Sequence Alignement aln <- pdbaln(files, outfile = tempfile())Reading PDB files: /tmp/RtmpTDihxb/split_chain/1a70_A.pdb /tmp/RtmpTDihxb/split_chain/1czp_A.pdb /tmp/RtmpTDihxb/split_chain/1frd_A.pdb /tmp/RtmpTDihxb/split_chain/1fxi_A.pdb /tmp/RtmpTDihxb/split_chain/1iue_A.pdb /tmp/RtmpTDihxb/split_chain/1pfd_A.pdb . PDB has ALT records, taking A only, rm.alt=TRUE ..... Extracting sequences pdb/seq: 1 name: /tmp/RtmpTDihxb/split_chain/1a70_A.pdb pdb/seq: 2 name: /tmp/RtmpTDihxb/split_chain/1czp_A.pdb PDB has ALT records, taking A only, rm.alt=TRUE pdb/seq: 3 name: /tmp/RtmpTDihxb/split_chain/1frd_A.pdb pdb/seq: 4 name: /tmp/RtmpTDihxb/split_chain/1fxi_A.pdb pdb/seq: 5 name: /tmp/RtmpTDihxb/split_chain/1iue_A.pdb pdb/seq: 6 name: /tmp/RtmpTDihxb/split_chain/1pfd_A.pdb## Read all pdb coordinates pdbs <- read.all(aln) ## Normal mode analysis on aligned data modes <- aanma(pdbs, rm.gaps=TRUE)Fitting pdb structuresdone Details of Scheduled Calculation: ... 6 input structures ... storing 282 eigenvectors for each structure ... dimension of x$U.subspace: ( 288x282x6 ) ... coordinate superposition prior to NM calculation ... aligned eigenvectors (gap containing positions removed) ... estimated memory usage of final 'eNMA' object: 3.7 Mb## Plot fluctuation data plot(modes, pdbs=pdbs)Extracting SSE from pdbs$sse attribute## Cluster on Fluctuation similariy sip <- sip(modes)
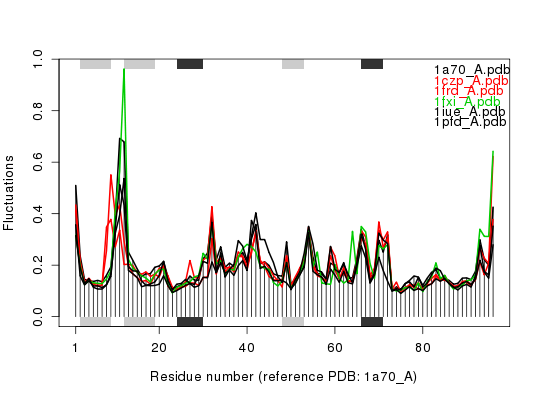
hc <- hclust(dist(sip)) col <- cutree(hc, k=3) ## Plot fluctuation data plot(modes, pdbs=pdbs, col=col)Extracting SSE from pdbs$sse attribute
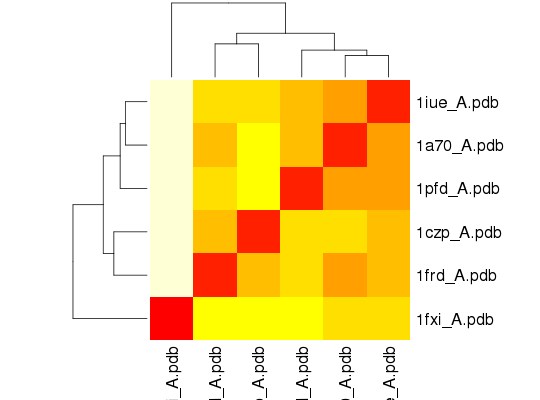
## RMSIP is pre-calculated heatmap(1-modes$rmsip)
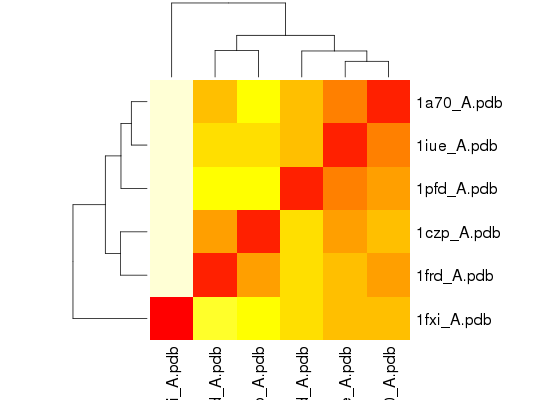
## Bhattacharyya coefficient bc <- bhattacharyya(modes)Calculating covariance matrices Calculating pairwise bhattacharyya coefsheatmap(1-bc)
See also
For normal mode analysis on single structure PDB:aanma
For conventional C-alpha based normal mode analysis:
nma, nma.pdbs.
For the analysis of the resulting ‘eNMA’ object:
mktrj.enma, dccm.enma,
plot.enma, cov.enma.
Similarity measures:
sip, covsoverlap,
bhattacharyya, rmsip.
Related functionality:
read.all.