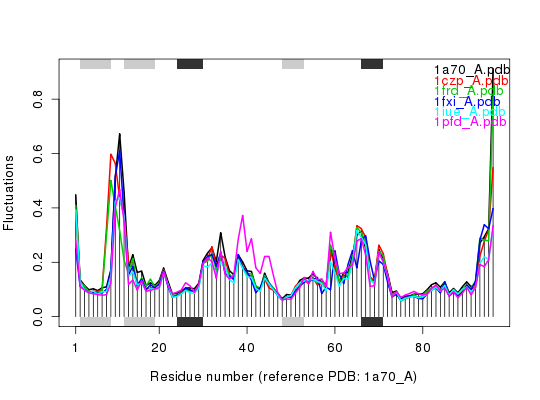
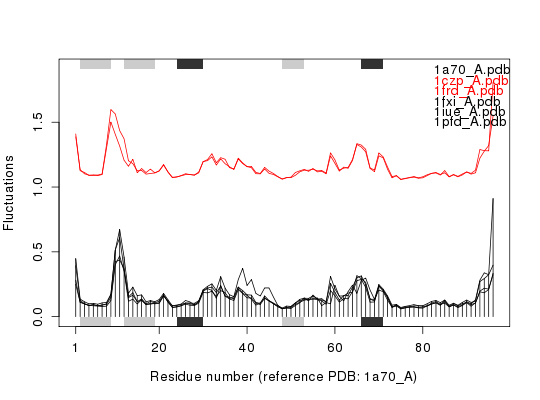
Plot eNMA Results
Usage
"plot"(x, pdbs = NULL, xlab = NULL, ylab="Fluctuations", ...)
Arguments
- x
- the results of ensemble NMA obtained with
nma.pdbs. Alternatively, a matrix in the similar format asenma$fluctuationscan be provided. - pdbs
- an object of class ‘pdbs’ in which the
‘enma’ object
xwas obtained from. If provided SSE data of the first structure ofpdbswill drawn. - xlab
- a label for the x axis.
- ylab
- labels for the y axes.
- ...
- extra plotting arguments passed to
plot.fluctthat effect the atomic fluctuations plot only.
Description
Produces a plot of atomic fluctuations of aligned normal modes.
Details
plot.enma produces a fluctuation plot of aligned nma
objects. If corresponding pdbs object is provided the plot
contains SSE annotation and appropriate resiude index numbering.
Value
-
Called for its effect.
References
Skjaerven, L. et al. (2014) BMC Bioinformatics 15, 399. Grant, B.J. et al. (2006) Bioinformatics 22, 2695--2696.
Examples
ids <- c("1a70_A", "1czp_A", "1frd_A", "1fxi_A", "1iue_A", "1pfd_A") raw.files <- get.pdb(ids, path = "raw_pdbs")Warning message: raw_pdbs/1a70.pdb exists. Skipping download Warning message: raw_pdbs/1czp.pdb exists. Skipping download Warning message: raw_pdbs/1frd.pdb exists. Skipping download Warning message: raw_pdbs/1fxi.pdb exists. Skipping download Warning message: raw_pdbs/1iue.pdb exists. Skipping download Warning message: raw_pdbs/1pfd.pdb exists. Skipping downloadfiles <- pdbsplit(raw.files, ids, path = "raw_pdbs/split_chain")|======================================================================| 100%## Sequence/structure alignement pdbs <- pdbaln(files)Reading PDB files: raw_pdbs/split_chain/1a70_A.pdb raw_pdbs/split_chain/1czp_A.pdb raw_pdbs/split_chain/1frd_A.pdb raw_pdbs/split_chain/1fxi_A.pdb raw_pdbs/split_chain/1iue_A.pdb raw_pdbs/split_chain/1pfd_A.pdb . PDB has ALT records, taking A only, rm.alt=TRUE ..... Extracting sequences pdb/seq: 1 name: raw_pdbs/split_chain/1a70_A.pdb pdb/seq: 2 name: raw_pdbs/split_chain/1czp_A.pdb PDB has ALT records, taking A only, rm.alt=TRUE pdb/seq: 3 name: raw_pdbs/split_chain/1frd_A.pdb pdb/seq: 4 name: raw_pdbs/split_chain/1fxi_A.pdb pdb/seq: 5 name: raw_pdbs/split_chain/1iue_A.pdb pdb/seq: 6 name: raw_pdbs/split_chain/1pfd_A.pdb## Normal mode analysis on aligned data modes <- nma(pdbs)Details of Scheduled Calculation: ... 6 input structures ... storing 282 eigenvectors for each structure ... dimension of x$U.subspace: ( 288x282x6 ) ... coordinate superposition prior to NM calculation ... aligned eigenvectors (gap containing positions removed) ... estimated memory usage of final 'eNMA' object: 3.7 Mb |======================================================================| 100%## Plot fluctuations plot(modes, pdbs=pdbs)Extracting SSE from pdbs$sse attribute## Group and spread fluctuation profiles hc <- hclust(as.dist(1-modes$rmsip))
col <- cutree(hc, k=2) plot(modes, pdbs=pdbs, col=col, spread=TRUE)Extracting SSE from pdbs$sse attribute