Prune A cna Network Object
Usage
prune.cna(x, edges.min = 1, size.min = 1)
Arguments
- x
- A protein network graph object as obtained from the ‘cna’ function.
- edges.min
- A single element numeric vector specifying the minimum number of edges that retained nodes should have. Nodes with less than ‘edges.min’ will be pruned.
- size.min
- A single element numeric vector specifying the minimum node size that retained nodes should have. Nodes with less composite residues than ‘size.min’ will be pruned.
Description
Remove nodes and their associated edges from a cna network graph.
Details
This function is useful for cleaning up cna network plots by removing, for example, small isolated nodes. The output is a new cna object minus the pruned nodes and their associated edges. Node naming is preserved.
Value
-
A cna class object, see function
cna for details.
References
Grant, B.J. et al. (2006) Bioinformatics 22, 2695--2696.
Note
Some improvements to this function are required, including a better effort to preserve the original community structure rather than calculating a new one. Also may consider removing nodes form the raw.network object that is returned also.
Examples
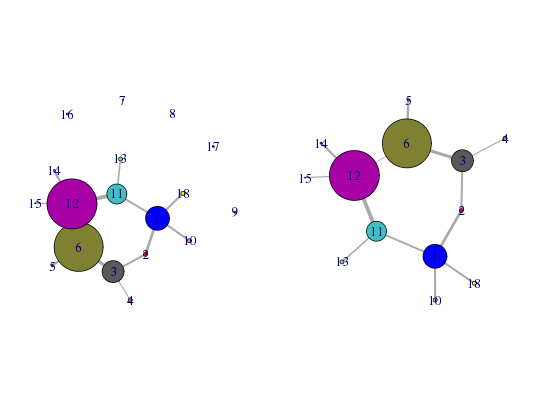
# Load the correlation network attach(hivp) # Read the starting PDB file to determine atom correspondence pdbfile <- system.file("examples/hivp.pdb", package="bio3d") pdb <- read.pdb(pdbfile) # Plot coarse grain network based on dynamically coupled communities par(mfcol=c(1,2), mar=c(0,0,0,0)) plot.cna(net)Obtaining estimated layout with fruchterman.reingold# Prune network dnet <- prune.cna(net, edges.min = 1)id size members 1 24 c(1:4, 94:107, 193:198) 2 4 5:8 3 22 c(9:23, 64:70) 4 4 24:27 5 2 28:29 6 49 c(30:63, 71:85) 7 1 86 8 1 87 9 2 88:89 10 4 90:93 11 20 c(108:113, 117:122, 163:170) 12 50 c(114:116, 129:162, 171:183) 13 4 123:126 14 2 127:128 15 1 184 16 2 185:186 17 2 187:188 18 4 189:192 Removing Nodes: 7, 8, 9, 16, 17 id size edges members 7 1 0 86 8 1 0 87 9 2 0 88:89 16 2 0 185:186 17 2 0 187:188plot(dnet)Obtaining estimated layout with fruchterman.reingold
detach(hivp)