Principal Component Analysis
Usage
"pca"(xyz, subset = rep(TRUE, nrow(as.matrix(xyz))), use.svd = FALSE, rm.gaps=FALSE, mass = NULL, ...)"print"(x, nmodes=6, ...)
Arguments
- xyz
- numeric matrix of Cartesian coordinates with a row per structure.
- subset
- an optional vector of numeric indices that selects a
subset of rows (e.g. experimental structures vs molecular dynamics
trajectory structures) from the full
xyzmatrix. Note: the fullxyzis projected onto this subspace. - use.svd
- logical, if TRUE singular value decomposition (SVD) is called instead of eigenvalue decomposition.
- rm.gaps
- logical, if TRUE gap positions (with missing coordinate data in any input structure) are removed before calculation. This is equivalent to removing NA cols from xyz.
- x
- an object of class
pca, as obtained from functionpca.xyz. - nmodes
- numeric, number of modes to be printed.
- mass
- a ‘pdb’ object or numeric vector of residue/atom masses.
By default (
mass=NULL), mass is ignored. If provided with a ‘pdb’ object, masses of all amino acids obtained fromaa2massare used. - ...
- additional arguments to
fit.xyz(forpca.xyz) or toprint(forprint.pca).
Description
Performs principal components analysis (PCA) on a xyz
numeric data matrix.
Note
If mass is provided, mass weighted coordinates will be considered,
and iteration of fitting onto the mean structure is performed internally.
The extra fitting process is to remove external translation and rotation
of the whole system. With this option, a direct comparison can be made
between PCs from pca.xyz and vibrational modes from
nma.pdb, with the fact that
A=k[B]TF^-1, where
A is the variance-covariance matrix, F the Hessian
matrix, k[B] the Boltzmann's constant, and T the
temperature.
Value
-
Returns a list with the following components:
- L
- eigenvalues.
- U
- eigenvectors (i.e. the x, y, and z variable loadings).
- z
- scores of the supplied
xyzon the pcs. - au
- atom-wise loadings (i.e. xyz normalized eigenvectors).
- sdev
- the standard deviations of the pcs.
- mean
- the means that were subtracted.
References
Grant, B.J. et al. (2006) Bioinformatics 22, 2695--2696.
Examples
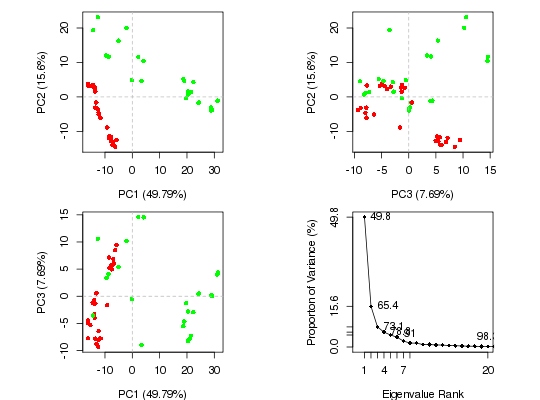
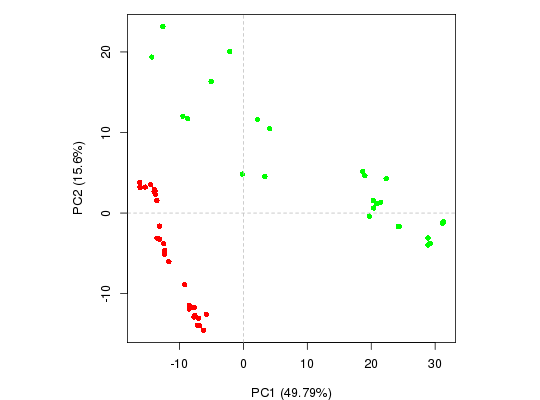
#-- Read transducin alignment and structures aln <- read.fasta(system.file("examples/transducin.fa",package="bio3d")) pdbs <- read.fasta.pdb(aln)pdb/seq: 1 name: http://www.rcsb.org/pdb/files/1TND.pdb pdb/seq: 2 name: http://www.rcsb.org/pdb/files/1TAD.pdb pdb/seq: 3 name: http://www.rcsb.org/pdb/files/1TAG.pdb pdb/seq: 4 name: http://www.rcsb.org/pdb/files/3V00.pdb pdb/seq: 5 name: http://www.rcsb.org/pdb/files/1FQJ.pdb pdb/seq: 6 name: http://www.rcsb.org/pdb/files/1FQK.pdb pdb/seq: 7 name: http://www.rcsb.org/pdb/files/2XNS.pdb PDB has ALT records, taking A only, rm.alt=TRUE pdb/seq: 8 name: http://www.rcsb.org/pdb/files/1KJY.pdb pdb/seq: 9 name: http://www.rcsb.org/pdb/files/2OM2.pdb pdb/seq: 10 name: http://www.rcsb.org/pdb/files/4G5Q.pdb PDB has ALT records, taking A only, rm.alt=TRUE pdb/seq: 11 name: http://www.rcsb.org/pdb/files/1GP2.pdb pdb/seq: 12 name: http://www.rcsb.org/pdb/files/1AGR.pdb pdb/seq: 13 name: http://www.rcsb.org/pdb/files/1CIP.pdb pdb/seq: 14 name: http://www.rcsb.org/pdb/files/1GFI.pdb pdb/seq: 15 name: http://www.rcsb.org/pdb/files/1GIA.pdb pdb/seq: 16 name: http://www.rcsb.org/pdb/files/2ZJY.pdb pdb/seq: 17 name: http://www.rcsb.org/pdb/files/3ONW.pdb pdb/seq: 18 name: http://www.rcsb.org/pdb/files/1BH2.pdb pdb/seq: 19 name: http://www.rcsb.org/pdb/files/1GG2.pdb pdb/seq: 20 name: http://www.rcsb.org/pdb/files/1GIT.pdb pdb/seq: 21 name: http://www.rcsb.org/pdb/files/3QI2.pdb pdb/seq: 22 name: http://www.rcsb.org/pdb/files/1SVK.pdb PDB has ALT records, taking A only, rm.alt=TRUE pdb/seq: 23 name: http://www.rcsb.org/pdb/files/1SVS.pdb pdb/seq: 24 name: http://www.rcsb.org/pdb/files/3FFA.pdb pdb/seq: 25 name: http://www.rcsb.org/pdb/files/1GIL.pdb pdb/seq: 26 name: http://www.rcsb.org/pdb/files/1AS0.pdb pdb/seq: 27 name: http://www.rcsb.org/pdb/files/1AS2.pdb pdb/seq: 28 name: http://www.rcsb.org/pdb/files/2ODE.pdb PDB has ALT records, taking A only, rm.alt=TRUE pdb/seq: 29 name: http://www.rcsb.org/pdb/files/2V4Z.pdb pdb/seq: 30 name: http://www.rcsb.org/pdb/files/4G5R.pdb pdb/seq: 31 name: http://www.rcsb.org/pdb/files/2IHB.pdb pdb/seq: 32 name: http://www.rcsb.org/pdb/files/4G5O.pdb PDB has ALT records, taking A only, rm.alt=TRUE# Find core core <- core.find(pdbs, #write.pdbs = TRUE, verbose=TRUE)core size 309 of 310 vol = 230.266 core size 308 of 310 vol = 205.404 core size 307 of 310 vol = 181.373 core size 306 of 310 vol = 160.715 core size 305 of 310 vol = 144.016 core size 304 of 310 vol = 129.352 core size 303 of 310 vol = 114.967 core size 302 of 310 vol = 104.141 core size 301 of 310 vol = 94.073 core size 300 of 310 vol = 86.326 core size 299 of 310 vol = 79.204 core size 298 of 310 vol = 73.164 core size 297 of 310 vol = 68.57 core size 296 of 310 vol = 64.346 core size 295 of 310 vol = 60.377 core size 294 of 310 vol = 56.493 core size 293 of 310 vol = 53.417 core size 292 of 310 vol = 50.379 core size 291 of 310 vol = 47.848 core size 290 of 310 vol = 45.441 core size 289 of 310 vol = 43.448 core size 288 of 310 vol = 41.464 core size 287 of 310 vol = 39.539 core size 286 of 310 vol = 37.874 core size 285 of 310 vol = 36.48 core size 284 of 310 vol = 35.105 core size 283 of 310 vol = 33.999 core size 282 of 310 vol = 32.973 core size 281 of 310 vol = 32.221 core size 280 of 310 vol = 31.472 core size 279 of 310 vol = 30.741 core size 278 of 310 vol = 30.032 core size 277 of 310 vol = 29.33 core size 276 of 310 vol = 28.694 core size 275 of 310 vol = 28.118 core size 274 of 310 vol = 27.536 core size 273 of 310 vol = 27.026 core size 272 of 310 vol = 26.523 core size 271 of 310 vol = 25.986 core size 270 of 310 vol = 25.44 core size 269 of 310 vol = 24.946 core size 268 of 310 vol = 24.593 core size 267 of 310 vol = 24.157 core size 266 of 310 vol = 23.721 core size 265 of 310 vol = 23.324 core size 264 of 310 vol = 22.977 core size 263 of 310 vol = 22.632 core size 262 of 310 vol = 22.297 core size 261 of 310 vol = 21.952 core size 260 of 310 vol = 21.581 core size 259 of 310 vol = 21.241 core size 258 of 310 vol = 20.864 core size 257 of 310 vol = 20.485 core size 256 of 310 vol = 20.143 core size 255 of 310 vol = 19.802 core size 254 of 310 vol = 19.453 core size 253 of 310 vol = 19.101 core size 252 of 310 vol = 18.859 core size 251 of 310 vol = 18.573 core size 250 of 310 vol = 18.271 core size 249 of 310 vol = 17.984 core size 248 of 310 vol = 17.686 core size 247 of 310 vol = 17.371 core size 246 of 310 vol = 17.089 core size 245 of 310 vol = 16.806 core size 244 of 310 vol = 16.535 core size 243 of 310 vol = 16.269 core size 242 of 310 vol = 16.035 core size 241 of 310 vol = 15.806 core size 240 of 310 vol = 15.549 core size 239 of 310 vol = 15.324 core size 238 of 310 vol = 15.102 core size 237 of 310 vol = 14.85 core size 236 of 310 vol = 14.638 core size 235 of 310 vol = 14.422 core size 234 of 310 vol = 14.24 core size 233 of 310 vol = 14.062 core size 232 of 310 vol = 13.858 core size 231 of 310 vol = 13.664 core size 230 of 310 vol = 13.472 core size 229 of 310 vol = 13.274 core size 228 of 310 vol = 13.101 core size 227 of 310 vol = 12.924 core size 226 of 310 vol = 12.736 core size 225 of 310 vol = 12.572 core size 224 of 310 vol = 12.373 core size 223 of 310 vol = 12.197 core size 222 of 310 vol = 12.049 core size 221 of 310 vol = 11.879 core size 220 of 310 vol = 11.673 core size 219 of 310 vol = 11.51 core size 218 of 310 vol = 11.381 core size 217 of 310 vol = 11.196 core size 216 of 310 vol = 11.037 core size 215 of 310 vol = 10.907 core size 214 of 310 vol = 10.765 core size 213 of 310 vol = 10.609 core size 212 of 310 vol = 10.466 core size 211 of 310 vol = 10.327 core size 210 of 310 vol = 10.184 core size 209 of 310 vol = 10.047 core size 208 of 310 vol = 9.896 core size 207 of 310 vol = 9.761 core size 206 of 310 vol = 9.652 core size 205 of 310 vol = 9.523 core size 204 of 310 vol = 9.378 core size 203 of 310 vol = 9.253 core size 202 of 310 vol = 9.159 core size 201 of 310 vol = 9.049 core size 200 of 310 vol = 8.929 core size 199 of 310 vol = 8.799 core size 198 of 310 vol = 8.669 core size 197 of 310 vol = 8.547 core size 196 of 310 vol = 8.41 core size 195 of 310 vol = 8.266 core size 194 of 310 vol = 8.127 core size 193 of 310 vol = 7.991 core size 192 of 310 vol = 7.854 core size 191 of 310 vol = 7.719 core size 190 of 310 vol = 7.59 core size 189 of 310 vol = 7.461 core size 188 of 310 vol = 7.336 core size 187 of 310 vol = 7.217 core size 186 of 310 vol = 7.087 core size 185 of 310 vol = 6.958 core size 184 of 310 vol = 6.842 core size 183 of 310 vol = 6.709 core size 182 of 310 vol = 6.581 core size 181 of 310 vol = 6.456 core size 180 of 310 vol = 6.336 core size 179 of 310 vol = 6.227 core size 178 of 310 vol = 6.129 core size 177 of 310 vol = 6.016 core size 176 of 310 vol = 5.934 core size 175 of 310 vol = 5.847 core size 174 of 310 vol = 5.764 core size 173 of 310 vol = 5.678 core size 172 of 310 vol = 5.6 core size 171 of 310 vol = 5.543 core size 170 of 310 vol = 5.461 core size 169 of 310 vol = 5.374 core size 168 of 310 vol = 5.291 core size 167 of 310 vol = 5.211 core size 166 of 310 vol = 5.133 core size 165 of 310 vol = 5.055 core size 164 of 310 vol = 4.975 core size 163 of 310 vol = 4.891 core size 162 of 310 vol = 4.801 core size 161 of 310 vol = 4.709 core size 160 of 310 vol = 4.644 core size 159 of 310 vol = 4.569 core size 158 of 310 vol = 4.479 core size 157 of 310 vol = 4.408 core size 156 of 310 vol = 4.343 core size 155 of 310 vol = 4.275 core size 154 of 310 vol = 4.191 core size 153 of 310 vol = 4.103 core size 152 of 310 vol = 4.016 core size 151 of 310 vol = 3.936 core size 150 of 310 vol = 3.864 core size 149 of 310 vol = 3.777 core size 148 of 310 vol = 3.687 core size 147 of 310 vol = 3.599 core size 146 of 310 vol = 3.514 core size 145 of 310 vol = 3.434 core size 144 of 310 vol = 3.349 core size 143 of 310 vol = 3.259 core size 142 of 310 vol = 3.18 core size 141 of 310 vol = 3.097 core size 140 of 310 vol = 3.025 core size 139 of 310 vol = 2.952 core size 138 of 310 vol = 2.875 core size 137 of 310 vol = 2.812 core size 136 of 310 vol = 2.729 core size 135 of 310 vol = 2.647 core size 134 of 310 vol = 2.563 core size 133 of 310 vol = 2.489 core size 132 of 310 vol = 2.407 core size 131 of 310 vol = 2.326 core size 130 of 310 vol = 2.26 core size 129 of 310 vol = 2.182 core size 128 of 310 vol = 2.109 core size 127 of 310 vol = 2.051 core size 126 of 310 vol = 1.993 core size 125 of 310 vol = 1.939 core size 124 of 310 vol = 1.883 core size 123 of 310 vol = 1.826 core size 122 of 310 vol = 1.772 core size 121 of 310 vol = 1.716 core size 120 of 310 vol = 1.672 core size 119 of 310 vol = 1.624 core size 118 of 310 vol = 1.581 core size 117 of 310 vol = 1.541 core size 116 of 310 vol = 1.508 core size 115 of 310 vol = 1.465 core size 114 of 310 vol = 1.425 core size 113 of 310 vol = 1.385 core size 112 of 310 vol = 1.35 core size 111 of 310 vol = 1.312 core size 110 of 310 vol = 1.277 core size 109 of 310 vol = 1.248 core size 108 of 310 vol = 1.22 core size 107 of 310 vol = 1.192 core size 106 of 310 vol = 1.159 core size 105 of 310 vol = 1.127 core size 104 of 310 vol = 1.099 core size 103 of 310 vol = 1.071 core size 102 of 310 vol = 1.043 core size 101 of 310 vol = 1.017 core size 100 of 310 vol = 0.993 core size 99 of 310 vol = 0.966 core size 98 of 310 vol = 0.945 core size 97 of 310 vol = 0.921 core size 96 of 310 vol = 0.9 core size 95 of 310 vol = 0.877 core size 94 of 310 vol = 0.854 core size 93 of 310 vol = 0.834 core size 92 of 310 vol = 0.813 core size 91 of 310 vol = 0.793 core size 90 of 310 vol = 0.772 core size 89 of 310 vol = 0.75 core size 88 of 310 vol = 0.73 core size 87 of 310 vol = 0.709 core size 86 of 310 vol = 0.69 core size 85 of 310 vol = 0.671 core size 84 of 310 vol = 0.652 core size 83 of 310 vol = 0.632 core size 82 of 310 vol = 0.613 core size 81 of 310 vol = 0.597 core size 80 of 310 vol = 0.581 core size 79 of 310 vol = 0.565 core size 78 of 310 vol = 0.547 core size 77 of 310 vol = 0.528 core size 76 of 310 vol = 0.512 core size 75 of 310 vol = 0.495 FINISHED: Min vol ( 0.5 ) reachedrm(list=c("pdbs", "core")) #-- OR for demo purposes just read previously saved transducin data attach(transducin) # Previously fitted coordinates based on sub 1.0A^3 core. See core.find() function. xyz <- pdbs$xyz #-- Do PCA ignoring gap containing positions pc.xray <- pca.xyz(xyz, rm.gaps=TRUE)NOTE: Removing 49 gap positions with missing coordinate data retaining 305 non-gap positions for analysis.# Plot results (conformer plots & scree plot overview) plot(pc.xray, col=annotation[, "color"]) # Plot a single conformer plot of PC1 v PC2 plot(pc.xray, pc.axes=1:2, col=annotation[, "color"])
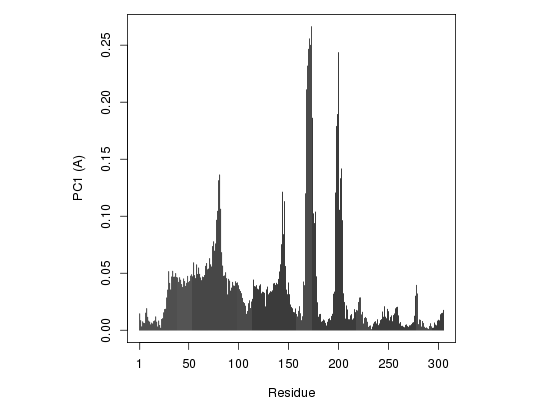
## Plot atom wise loadings plot.bio3d(pc.xray$au[,1], ylab="PC1 (A)")
# PDB server connection required - testing excluded ## Plot loadings in relation to reference structure 1TAG pdb <- read.pdb("1tag")Note: Accessing on-line PDB fileind <- grep("1TAG", pdbs$id) ## location in alignment resno <- pdbs$resno[ind, !is.gap(pdbs)] ## non-gap residues tpdb <- trim.pdb(pdb, resno=resno) op <- par(no.readonly=TRUE) par(mfrow = c(3, 1), cex = 0.6, mar = c(3, 4, 1, 1)) plot.bio3d(pc.xray$au[,1], resno, ylab="PC1 (A)", sse=tpdb) plot.bio3d(pc.xray$au[,2], resno, ylab="PC2 (A)", sse=tpdb) plot.bio3d(pc.xray$au[,3], resno, ylab="PC3 (A)", sse=tpdb)
par(op) # Write PC trajectory resno = pdbs$resno[1, !is.gap(pdbs)] resid = aa123(pdbs$ali[1, !is.gap(pdbs)]) a <- mktrj.pca(pc.xray, pc=1, file="pc1.pdb", resno=resno, resid=resid ) b <- mktrj.pca(pc.xray, pc=2, file="pc2.pdb", resno=resno, resid=resid ) c <- mktrj.pca(pc.xray, pc=3, file="pc3.pdb", resno=resno, resid=resid ) detach(transducin)