Principal Component Analysis
Usage
"pca"(data, ...)
Arguments
- data
- numeric matrix of torsion angles with a row per structure.
- ...
- additional arguments passed to the method
pca.xyz.
Description
Performs principal components analysis (PCA) on torsion angle data.
Value
-
Returns a list with the following components:
- L
- eigenvalues.
- U
- eigenvectors (i.e. the variable loadings).
- z.u
- scores of the supplied
dataon the pcs. - sdev
- the standard deviations of the pcs.
- mean
- the means that were subtracted.
References
Grant, B.J. et al. (2006) Bioinformatics 22, 2695--2696.
Examples
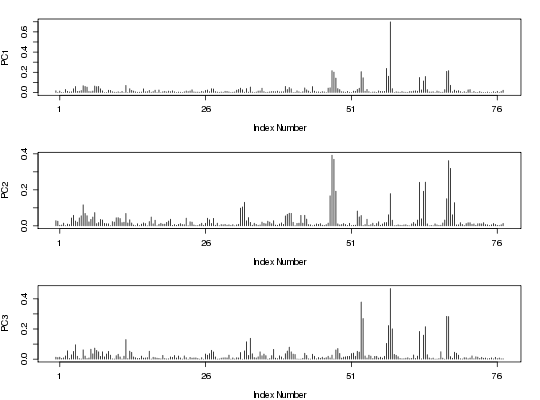
##-- PCA on torsion data for multiple PDBs attach(kinesin) gaps.pos <- gap.inspect(pdbs$xyz) tor <- t(apply( pdbs$xyz[, gaps.pos$f.inds], 1, torsion.xyz, atm.inc=1)) pc.tor <- pca.tor(tor[,-c(1,233,234,235)]) #plot(pc.tor) plot.pca.loadings(pc.tor) detach(kinesin)
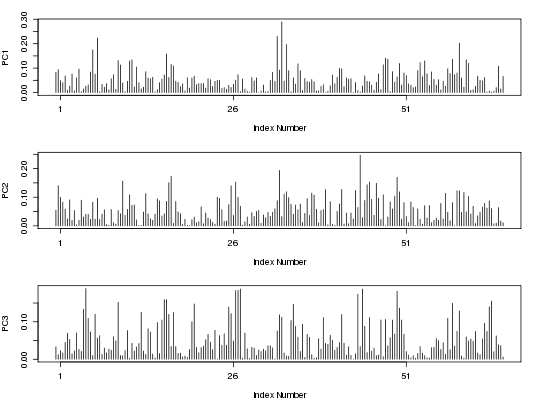
##-- PCA on torsion data from an MD trajectory trj <- read.dcd( system.file("examples/hivp.dcd", package="bio3d") )NATOM = 198 NFRAME= 117 ISTART= 0 last = 117 nstep = 117 nfile = 117 NSAVE = 1 NDEGF = 0 version 24 Reading (x100) |======================================================================| 100%tor <- t(apply(trj, 1, torsion.xyz, atm.inc=1)) gaps <- gap.inspect(tor) pc.tor <- pca.tor(tor[,gaps$f.inds]) plot.pca.loadings(pc.tor)