GeoStaS Domain Finder
Usage
geostas(...)"geostas"(...)"geostas"(xyz, amsm = NULL, k = 3, pairwise = TRUE, clustalg = "kmeans", fit = TRUE, ncore = NULL, verbose=TRUE, ...)"geostas"(nma, m.inds = 7:11, verbose=TRUE, ...)"geostas"(enma, pdbs = NULL, m.inds = 1:5, verbose=TRUE, ...)"geostas"(pdb, inds = NULL, verbose=TRUE, ...)"geostas"(pdbs, verbose=TRUE, ...)amsm.xyz(xyz, ncore = NULL)"print"(x, ...)
Arguments
- ...
- arguments passed to and from functions, such as
kmeans, andhclustwhich are called internally ingeostas.xyz. - xyz
- numeric matrix of xyz coordinates as obtained e.g. by
read.ncdf,read.dcd, ormktrj. - amsm
- a numeric matrix as obtained by
amsm.xyz(convenient e.g. for re-doing only the clustering analysis of the ‘AMSM’ matrix). - k
- an integer scalar or vector with the desired number of groups.
- pairwise
- logical, if TRUE use pairwise clustering of the atomic movement similarity matrix (AMSM), else columnwise.
- clustalg
- a character string specifing the clustering algorithm. Allowed values are ‘kmeans’ and ‘hclust’.
- fit
- logical, if TRUE coordinate superposition on identified core atoms is performed prior to the calculation of the AMS matrix.
- ncore
- number of CPU cores used to do the calculation.
ncore>1requires package ‘parallel’ installed. - verbose
- logical, if TRUE details of the geostas calculations are printed to screen.
- nma
- an ‘nma’ object as obtained from function
nma. Functionmktrjis used internally to generate a trajectory based on the normal modes. - m.inds
- the mode number(s) along which trajectory should be
made (see function
mktrj). - enma
- an ‘enma’ object as obtained from function
nma.pdbs. Functionmktrjis used internally to generate a trajectory based on the normal modes. - pdbs
- a ‘pdbs’ object as obtained from function
pdbalnorread.fasta.pdb. - pdb
- a ‘pdb’ object as obtained from function
read.pdb. - inds
- a ‘select’ object as obtained from function
atom.selectgiving the atomic indices at which the calculation should be based. By default the function will attempt to locate C-alpha atoms using functionatom.select. - x
- a ‘geostas’ object as obtained from function
geostas.
Description
Identifies geometrically stable domains in biomolecules
Details
This function attempts to identify rigid domains in a protein (or nucleic acid) structure based on an structural ensemble, e.g. obtained from NMR experiments, molecular dynamics simulations, or normal mode analysis.
The algorithm is based on a geometric approach for comparing pairwise
traces of atomic motion and the search for their best superposition
using a quaternion representation of rotation. The result is stored in
a NxN atomic movement similarity matrix (AMSM) describing the
correspondence between all pairs of atom motion. Rigid domains are
obtained by clustering the elements of the AMS matrix
(pairwise=TRUE), or alternatively, the columns similarity
(pairwise=FALSE), using either K-means (kmeans)
or hierarchical (hclust) clustering.
Compared to the conventional cross-correlation matrix (see function
dccm) the “geostas” approach provide
functionality to also detect domains involved in rotational
motions (i.e. two atoms located on opposite sides of a rotating
domain will appear as anti-correlated in the cross-correlation matrix,
but should obtain a high similarity coefficient in the AMS matrix).
See examples for more details.
Note
The current implementation in Bio3D uses a different fitting and clustering approach than the original Java implementation. The results will therefore differ.
Value
-
Returns a list object of type ‘geostas’ with the following components:
- amsm
- a numeric matrix of atomic movement similarity (AMSM).
- fit.inds
- a numeric vector of xyz indices used for fitting.
- grps
- a numeric vector containing the domain assignment per residue.
- atomgrps
- a numeric vector containing the domain assignment per
atom (only provided for
geostas.pdb). - inds
- a list of atom ‘select’ objects with indices to corresponding to the identified domains.
References
Romanowska, J. et al. (2012) JCTC 8, 2588--2599. Skjaerven, L. et al. (2014) BMC Bioinformatics 15, 399. Grant, B.J. et al. (2006) Bioinformatics 22, 2695--2696.
Examples
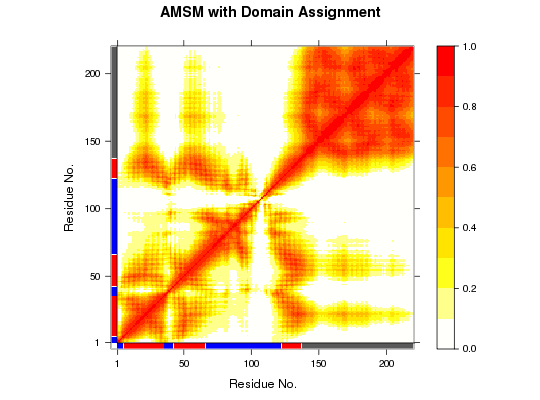
# PDB server connection required - testing excluded #### NMR-ensemble example ## Read a multi-model PDB file pdb <- read.pdb("1d1d", multi=TRUE)Note: Accessing on-line PDB file## Find domains and write PDB gs <- geostas(pdb, fit=TRUE).. 220 'calpha' atoms selected .. 'xyz' coordinate data with 20 frames .. 'fit=TRUE': running function 'core.find' .. coordinates are superimposed to core region .. calculating atomic movement similarity matrix ('amsm.xyz()') .. dimensions of AMSM are 220x220 .. clustering AMSM using 'kmeans' .. converting indices to match input 'pdb' object (additional attribute 'atomgrps' generated)## Plot a atomic movement similarity matrix plot.geostas(gs, contour=FALSE) ## Fit all frames to the 'first' domain domain.inds <- gs$inds[[1]]
xyz <- pdbfit(pdb, inds=domain.inds) #write.pdb(pdb, xyz=xyz, chain=gs$atomgrps) #### NMA example ## Fetch stucture pdb <- read.pdb("1crn")Note: Accessing on-line PDB file## Calculate (vibrational) normal modes modes <- nma(pdb)Building Hessian... Done in 0.013 seconds. Diagonalizing Hessian... Done in 0.009 seconds.## Find domains gs <- geostas(modes, k=2).. generating trajectory from 5 modes .. 'xyz' coordinate data with 85 frames .. 'fit=TRUE': running function 'core.find' .. coordinates are superimposed to core region .. calculating atomic movement similarity matrix ('amsm.xyz()') .. dimensions of AMSM are 46x46 .. clustering AMSM using 'kmeans'## Write NMA trajectory with domain assignment mktrj(modes, mode=7, chain=gs$grps) ## Redo geostas domain clustering gs <- geostas(modes, amsm=gs$amsm, k=5).. generating trajectory from 5 modes .. 'xyz' coordinate data with 85 frames .. coordinates are not superimposed prior to geostas calculation .. clustering AMSM using 'kmeans'#### Trajectory example ## Read inn DCD trajectory file, fit coordinates dcdfile <- system.file("examples/hivp.dcd", package = "bio3d") trj <- read.dcd(dcdfile)NATOM = 198 NFRAME= 117 ISTART= 0 last = 117 nstep = 117 nfile = 117 NSAVE = 1 NDEGF = 0 version 24 Reading (x100) |======================================================================| 100%xyz <- fit.xyz(trj[1,], trj)Warning message: No fitting indices provided, using the 198 non NA positions## Find domains gs <- geostas(xyz, k=3, fit=FALSE).. 'xyz' coordinate data with 117 frames .. coordinates are not superimposed prior to geostas calculation .. calculating atomic movement similarity matrix ('amsm.xyz()') .. dimensions of AMSM are 198x198 .. clustering AMSM using 'kmeans'## Principal component analysis pc.md <- pca.xyz(xyz) ## Visualize PCs with colored domains (chain ID) mktrj(pc.md, pc=1, chain=gs$grps) #### X-ray ensemble GroEL subunits # Define the ensemble PDB-ids ids <- c("1sx4_[A,B,H,I]", "1xck_[A-B]", "1sx3_[A-B]", "4ab3_[A-B]") # Download and split PDBs by chain ID raw.files <- get.pdb(ids, path = "raw_pdbs", gzip = TRUE) files <- pdbsplit(raw.files, ids, path = "raw_pdbs/split_chain/")|======================================================================| 100%# Align structures pdbs <- pdbaln(files)Reading PDB files: raw_pdbs/split_chain//1sx4_A.pdb raw_pdbs/split_chain//1sx4_B.pdb raw_pdbs/split_chain//1sx4_H.pdb raw_pdbs/split_chain//1sx4_I.pdb raw_pdbs/split_chain//1xck_A.pdb raw_pdbs/split_chain//1xck_B.pdb raw_pdbs/split_chain//1sx3_A.pdb raw_pdbs/split_chain//1sx3_B.pdb raw_pdbs/split_chain//4ab3_A.pdb raw_pdbs/split_chain//4ab3_B.pdb .......... Extracting sequences pdb/seq: 1 name: raw_pdbs/split_chain//1sx4_A.pdb pdb/seq: 2 name: raw_pdbs/split_chain//1sx4_B.pdb pdb/seq: 3 name: raw_pdbs/split_chain//1sx4_H.pdb pdb/seq: 4 name: raw_pdbs/split_chain//1sx4_I.pdb pdb/seq: 5 name: raw_pdbs/split_chain//1xck_A.pdb pdb/seq: 6 name: raw_pdbs/split_chain//1xck_B.pdb pdb/seq: 7 name: raw_pdbs/split_chain//1sx3_A.pdb pdb/seq: 8 name: raw_pdbs/split_chain//1sx3_B.pdb pdb/seq: 9 name: raw_pdbs/split_chain//4ab3_A.pdb pdb/seq: 10 name: raw_pdbs/split_chain//4ab3_B.pdb# Find domains gs <- geostas(pdbs, k=4, fit=TRUE).. 1569 non-gap positions selected .. 'xyz' coordinate data with 10 frames .. 'fit=TRUE': running function 'core.find' .. coordinates are superimposed to core region .. calculating atomic movement similarity matrix ('amsm.xyz()') .. dimensions of AMSM are 523x523 .. clustering AMSM using 'kmeans'# Superimpose to core region pdbs$xyz <- pdbfit(pdbs, inds=gs$fit.inds) # Principal component analysis pc.xray <- pca(pdbs) # Visualize PCs with colored domains (chain ID) mktrj(pc.xray, pc=1, chain=gs$grps) ##- Same, but more manual approach gaps.pos <- gap.inspect(pdbs$xyz) # Find core region core <- core.find(pdbs)core size 522 of 523 vol = 6446.395 core size 521 of 523 vol = 6316.869 core size 520 of 523 vol = 6192.665 core size 519 of 523 vol = 6071.24 core size 518 of 523 vol = 5956 core size 517 of 523 vol = 5841.265 core size 516 of 523 vol = 5724.389 core size 515 of 523 vol = 5598.024 core size 514 of 523 vol = 5505.679 core size 513 of 523 vol = 5420.778 core size 512 of 523 vol = 5331.605 core size 511 of 523 vol = 5248.446 core size 510 of 523 vol = 5168.015 core size 509 of 523 vol = 5097.58 core size 508 of 523 vol = 5011.538 core size 507 of 523 vol = 4923.883 core size 506 of 523 vol = 4844.813 core size 505 of 523 vol = 4776.429 core size 504 of 523 vol = 4692.108 core size 503 of 523 vol = 4618.873 core size 502 of 523 vol = 4542.239 core size 501 of 523 vol = 4473.649 core size 500 of 523 vol = 4403.837 core size 499 of 523 vol = 4334.207 core size 498 of 523 vol = 4286.785 core size 497 of 523 vol = 4223.922 core size 496 of 523 vol = 4178.361 core size 495 of 523 vol = 4114.812 core size 494 of 523 vol = 4065.855 core size 493 of 523 vol = 4021.027 core size 492 of 523 vol = 3977.844 core size 491 of 523 vol = 3927.871 core size 490 of 523 vol = 3879.635 core size 489 of 523 vol = 3827.791 core size 488 of 523 vol = 3771.895 core size 487 of 523 vol = 3720.998 core size 486 of 523 vol = 3671.378 core size 485 of 523 vol = 3613.118 core size 484 of 523 vol = 3560.233 core size 483 of 523 vol = 3505.806 core size 482 of 523 vol = 3452.731 core size 481 of 523 vol = 3414.454 core size 480 of 523 vol = 3375.548 core size 479 of 523 vol = 3329.392 core size 478 of 523 vol = 3285.413 core size 477 of 523 vol = 3244.743 core size 476 of 523 vol = 3201.697 core size 475 of 523 vol = 3162.417 core size 474 of 523 vol = 3123.15 core size 473 of 523 vol = 3079.758 core size 472 of 523 vol = 3036.888 core size 471 of 523 vol = 2987.262 core size 470 of 523 vol = 2957.669 core size 469 of 523 vol = 2923.79 core size 468 of 523 vol = 2889.149 core size 467 of 523 vol = 2856.689 core size 466 of 523 vol = 2818.157 core size 465 of 523 vol = 2784.161 core size 464 of 523 vol = 2745.535 core size 463 of 523 vol = 2702.435 core size 462 of 523 vol = 2651.363 core size 461 of 523 vol = 2622.559 core size 460 of 523 vol = 2578.404 core size 459 of 523 vol = 2550.123 core size 458 of 523 vol = 2511.64 core size 457 of 523 vol = 2476.22 core size 456 of 523 vol = 2453.882 core size 455 of 523 vol = 2428.977 core size 454 of 523 vol = 2406.363 core size 453 of 523 vol = 2379.14 core size 452 of 523 vol = 2346.55 core size 451 of 523 vol = 2306.998 core size 450 of 523 vol = 2278.307 core size 449 of 523 vol = 2257.135 core size 448 of 523 vol = 2234.657 core size 447 of 523 vol = 2210.146 core size 446 of 523 vol = 2192.121 core size 445 of 523 vol = 2177.246 core size 444 of 523 vol = 2153.257 core size 443 of 523 vol = 2127.202 core size 442 of 523 vol = 2104.921 core size 441 of 523 vol = 2079.106 core size 440 of 523 vol = 2043.655 core size 439 of 523 vol = 1996.65 core size 438 of 523 vol = 1957.765 core size 437 of 523 vol = 1931.91 core size 436 of 523 vol = 1908.683 core size 435 of 523 vol = 1892.177 core size 434 of 523 vol = 1875.47 core size 433 of 523 vol = 1850.336 core size 432 of 523 vol = 1829.666 core size 431 of 523 vol = 1808.438 core size 430 of 523 vol = 1790.485 core size 429 of 523 vol = 1767.266 core size 428 of 523 vol = 1755.76 core size 427 of 523 vol = 1739.14 core size 426 of 523 vol = 1716.73 core size 425 of 523 vol = 1680.031 core size 424 of 523 vol = 1653.883 core size 423 of 523 vol = 1636.674 core size 422 of 523 vol = 1610.199 core size 421 of 523 vol = 1589.881 core size 420 of 523 vol = 1563.268 core size 419 of 523 vol = 1555.686 core size 418 of 523 vol = 1525.146 core size 417 of 523 vol = 1505.498 core size 416 of 523 vol = 1480.181 core size 415 of 523 vol = 1466.243 core size 414 of 523 vol = 1442.466 core size 413 of 523 vol = 1413.597 core size 412 of 523 vol = 1392.795 core size 411 of 523 vol = 1377.501 core size 410 of 523 vol = 1353.087 core size 409 of 523 vol = 1310.301 core size 408 of 523 vol = 1271.485 core size 407 of 523 vol = 1251.26 core size 406 of 523 vol = 1233.928 core size 405 of 523 vol = 1214.185 core size 404 of 523 vol = 1203.452 core size 403 of 523 vol = 1180.312 core size 402 of 523 vol = 1153.446 core size 401 of 523 vol = 1133.862 core size 400 of 523 vol = 1112.512 core size 399 of 523 vol = 1086.741 core size 398 of 523 vol = 1076.38 core size 397 of 523 vol = 1051.229 core size 396 of 523 vol = 1036.102 core size 395 of 523 vol = 1013.196 core size 394 of 523 vol = 997.744 core size 393 of 523 vol = 970.507 core size 392 of 523 vol = 964.526 core size 391 of 523 vol = 953.432 core size 390 of 523 vol = 930.833 core size 389 of 523 vol = 906.683 core size 388 of 523 vol = 891.721 core size 387 of 523 vol = 893.406 core size 386 of 523 vol = 881.972 core size 385 of 523 vol = 863.539 core size 384 of 523 vol = 849.537 core size 383 of 523 vol = 824.13 core size 382 of 523 vol = 807.908 core size 381 of 523 vol = 788.814 core size 380 of 523 vol = 768.701 core size 379 of 523 vol = 758.089 core size 378 of 523 vol = 735.51 core size 377 of 523 vol = 714.724 core size 376 of 523 vol = 696.521 core size 375 of 523 vol = 682.95 core size 374 of 523 vol = 667.834 core size 373 of 523 vol = 649.667 core size 372 of 523 vol = 626.344 core size 371 of 523 vol = 608.313 core size 370 of 523 vol = 590.196 core size 369 of 523 vol = 575.976 core size 368 of 523 vol = 560.374 core size 367 of 523 vol = 543.547 core size 366 of 523 vol = 517.475 core size 365 of 523 vol = 496.901 core size 364 of 523 vol = 474.991 core size 363 of 523 vol = 456.949 core size 362 of 523 vol = 435.764 core size 361 of 523 vol = 417.323 core size 360 of 523 vol = 396.396 core size 359 of 523 vol = 385.105 core size 358 of 523 vol = 370.818 core size 357 of 523 vol = 352.351 core size 356 of 523 vol = 334.495 core size 355 of 523 vol = 313.975 core size 354 of 523 vol = 302.539 core size 353 of 523 vol = 285.208 core size 352 of 523 vol = 276.784 core size 351 of 523 vol = 261.112 core size 350 of 523 vol = 251.034 core size 349 of 523 vol = 235.907 core size 348 of 523 vol = 219.714 core size 347 of 523 vol = 208.271 core size 346 of 523 vol = 196.145 core size 345 of 523 vol = 185.944 core size 344 of 523 vol = 177.86 core size 343 of 523 vol = 171.468 core size 342 of 523 vol = 164.953 core size 341 of 523 vol = 159.763 core size 340 of 523 vol = 154.583 core size 339 of 523 vol = 150.647 core size 338 of 523 vol = 144.884 core size 337 of 523 vol = 140.682 core size 336 of 523 vol = 137.392 core size 335 of 523 vol = 135.289 core size 334 of 523 vol = 132.499 core size 333 of 523 vol = 127.929 core size 332 of 523 vol = 125.773 core size 331 of 523 vol = 123.115 core size 330 of 523 vol = 120.798 core size 329 of 523 vol = 118.347 core size 328 of 523 vol = 116.495 core size 327 of 523 vol = 114.297 core size 326 of 523 vol = 111.803 core size 325 of 523 vol = 109.59 core size 324 of 523 vol = 107.045 core size 323 of 523 vol = 105.696 core size 322 of 523 vol = 103.604 core size 321 of 523 vol = 101.472 core size 320 of 523 vol = 99.945 core size 319 of 523 vol = 98.011 core size 318 of 523 vol = 96.056 core size 317 of 523 vol = 94.061 core size 316 of 523 vol = 92.085 core size 315 of 523 vol = 90.631 core size 314 of 523 vol = 88.889 core size 313 of 523 vol = 86.794 core size 312 of 523 vol = 85.164 core size 311 of 523 vol = 83.756 core size 310 of 523 vol = 82.212 core size 309 of 523 vol = 80.491 core size 308 of 523 vol = 79.432 core size 307 of 523 vol = 77.712 core size 306 of 523 vol = 76.762 core size 305 of 523 vol = 75.416 core size 304 of 523 vol = 74.236 core size 303 of 523 vol = 72.55 core size 302 of 523 vol = 70.953 core size 301 of 523 vol = 69.694 core size 300 of 523 vol = 68.773 core size 299 of 523 vol = 67.108 core size 298 of 523 vol = 65.748 core size 297 of 523 vol = 64.121 core size 296 of 523 vol = 62.62 core size 295 of 523 vol = 61.392 core size 294 of 523 vol = 60.138 core size 293 of 523 vol = 58.829 core size 292 of 523 vol = 57.482 core size 291 of 523 vol = 56.099 core size 290 of 523 vol = 54.915 core size 289 of 523 vol = 53.46 core size 288 of 523 vol = 52.082 core size 287 of 523 vol = 50.707 core size 286 of 523 vol = 49.531 core size 285 of 523 vol = 48.307 core size 284 of 523 vol = 47.016 core size 283 of 523 vol = 45.845 core size 282 of 523 vol = 44.418 core size 281 of 523 vol = 42.962 core size 280 of 523 vol = 41.669 core size 279 of 523 vol = 40.049 core size 278 of 523 vol = 38.45 core size 277 of 523 vol = 37.089 core size 276 of 523 vol = 35.634 core size 275 of 523 vol = 34.139 core size 274 of 523 vol = 32.483 core size 273 of 523 vol = 31.107 core size 272 of 523 vol = 29.88 core size 271 of 523 vol = 28.715 core size 270 of 523 vol = 27.409 core size 269 of 523 vol = 26.127 core size 268 of 523 vol = 24.934 core size 267 of 523 vol = 23.767 core size 266 of 523 vol = 22.672 core size 265 of 523 vol = 21.634 core size 264 of 523 vol = 20.776 core size 263 of 523 vol = 19.848 core size 262 of 523 vol = 19.073 core size 261 of 523 vol = 18.17 core size 260 of 523 vol = 17.308 core size 259 of 523 vol = 16.537 core size 258 of 523 vol = 15.811 core size 257 of 523 vol = 15.148 core size 256 of 523 vol = 14.55 core size 255 of 523 vol = 14.004 core size 254 of 523 vol = 13.54 core size 253 of 523 vol = 13.076 core size 252 of 523 vol = 12.704 core size 251 of 523 vol = 12.291 core size 250 of 523 vol = 11.884 core size 249 of 523 vol = 11.514 core size 248 of 523 vol = 11.139 core size 247 of 523 vol = 10.79 core size 246 of 523 vol = 10.417 core size 245 of 523 vol = 10.002 core size 244 of 523 vol = 9.618 core size 243 of 523 vol = 9.302 core size 242 of 523 vol = 9.032 core size 241 of 523 vol = 8.772 core size 240 of 523 vol = 8.521 core size 239 of 523 vol = 8.265 core size 238 of 523 vol = 7.983 core size 237 of 523 vol = 7.677 core size 236 of 523 vol = 7.419 core size 235 of 523 vol = 7.209 core size 234 of 523 vol = 6.977 core size 233 of 523 vol = 6.757 core size 232 of 523 vol = 6.592 core size 231 of 523 vol = 6.412 core size 230 of 523 vol = 6.224 core size 229 of 523 vol = 6.015 core size 228 of 523 vol = 5.838 core size 227 of 523 vol = 5.631 core size 226 of 523 vol = 5.426 core size 225 of 523 vol = 5.269 core size 224 of 523 vol = 5.08 core size 223 of 523 vol = 4.887 core size 222 of 523 vol = 4.715 core size 221 of 523 vol = 4.537 core size 220 of 523 vol = 4.375 core size 219 of 523 vol = 4.222 core size 218 of 523 vol = 4.087 core size 217 of 523 vol = 3.963 core size 216 of 523 vol = 3.867 core size 215 of 523 vol = 3.763 core size 214 of 523 vol = 3.638 core size 213 of 523 vol = 3.534 core size 212 of 523 vol = 3.422 core size 211 of 523 vol = 3.343 core size 210 of 523 vol = 3.268 core size 209 of 523 vol = 3.173 core size 208 of 523 vol = 3.082 core size 207 of 523 vol = 2.993 core size 206 of 523 vol = 2.912 core size 205 of 523 vol = 2.84 core size 204 of 523 vol = 2.752 core size 203 of 523 vol = 2.679 core size 202 of 523 vol = 2.595 core size 201 of 523 vol = 2.539 core size 200 of 523 vol = 2.484 core size 199 of 523 vol = 2.43 core size 198 of 523 vol = 2.377 core size 197 of 523 vol = 2.33 core size 196 of 523 vol = 2.289 core size 195 of 523 vol = 2.247 core size 194 of 523 vol = 2.203 core size 193 of 523 vol = 2.158 core size 192 of 523 vol = 2.118 core size 191 of 523 vol = 2.075 core size 190 of 523 vol = 2.037 core size 189 of 523 vol = 1.999 core size 188 of 523 vol = 1.959 core size 187 of 523 vol = 1.919 core size 186 of 523 vol = 1.878 core size 185 of 523 vol = 1.844 core size 184 of 523 vol = 1.807 core size 183 of 523 vol = 1.77 core size 182 of 523 vol = 1.735 core size 181 of 523 vol = 1.708 core size 180 of 523 vol = 1.673 core size 179 of 523 vol = 1.644 core size 178 of 523 vol = 1.609 core size 177 of 523 vol = 1.577 core size 176 of 523 vol = 1.539 core size 175 of 523 vol = 1.52 core size 174 of 523 vol = 1.495 core size 173 of 523 vol = 1.471 core size 172 of 523 vol = 1.444 core size 171 of 523 vol = 1.419 core size 170 of 523 vol = 1.39 core size 169 of 523 vol = 1.368 core size 168 of 523 vol = 1.347 core size 167 of 523 vol = 1.324 core size 166 of 523 vol = 1.3 core size 165 of 523 vol = 1.279 core size 164 of 523 vol = 1.258 core size 163 of 523 vol = 1.234 core size 162 of 523 vol = 1.211 core size 161 of 523 vol = 1.182 core size 160 of 523 vol = 1.158 core size 159 of 523 vol = 1.132 core size 158 of 523 vol = 1.105 core size 157 of 523 vol = 1.082 core size 156 of 523 vol = 1.061 core size 155 of 523 vol = 1.041 core size 154 of 523 vol = 1.02 core size 153 of 523 vol = 1.001 core size 152 of 523 vol = 0.98 core size 151 of 523 vol = 0.949 core size 150 of 523 vol = 0.931 core size 149 of 523 vol = 0.914 core size 148 of 523 vol = 0.899 core size 147 of 523 vol = 0.88 core size 146 of 523 vol = 0.867 core size 145 of 523 vol = 0.847 core size 144 of 523 vol = 0.834 core size 143 of 523 vol = 0.814 core size 142 of 523 vol = 0.797 core size 141 of 523 vol = 0.778 core size 140 of 523 vol = 0.761 core size 139 of 523 vol = 0.746 core size 138 of 523 vol = 0.732 core size 137 of 523 vol = 0.72 core size 136 of 523 vol = 0.698 core size 135 of 523 vol = 0.686 core size 134 of 523 vol = 0.668 core size 133 of 523 vol = 0.654 core size 132 of 523 vol = 0.637 core size 131 of 523 vol = 0.626 core size 130 of 523 vol = 0.611 core size 129 of 523 vol = 0.598 core size 128 of 523 vol = 0.585 core size 127 of 523 vol = 0.57 core size 126 of 523 vol = 0.554 core size 125 of 523 vol = 0.541 core size 124 of 523 vol = 0.529 core size 123 of 523 vol = 0.513 core size 122 of 523 vol = 0.496 FINISHED: Min vol ( 0.5 ) reached# Fit to core region xyz <- fit.xyz(pdbs$xyz[1, gaps.pos$f.inds], pdbs$xyz[, gaps.pos$f.inds], fixed.inds=core$xyz, mobile.inds=core$xyz) # Find domains gs <- geostas(xyz, k=4, fit=FALSE).. 'xyz' coordinate data with 10 frames .. coordinates are not superimposed prior to geostas calculation .. calculating atomic movement similarity matrix ('amsm.xyz()') .. dimensions of AMSM are 523x523 .. clustering AMSM using 'kmeans'# Perform PCA pc.xray <- pca.xyz(xyz) # Make trajectory mktrj(pc.xray, pc=1, chain=gs$grps)