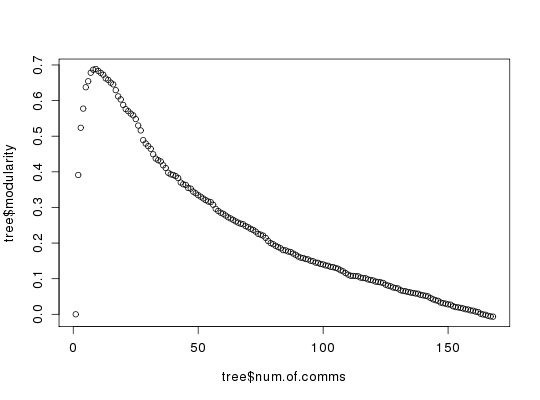
Reconstruction of the Girvan-Newman Community Tree for a CNA Class Object.
Usage
community.tree(x, rescale=FALSE)
Arguments
- x
- A protein network graph object as obtained from the ‘cna’ function.
- rescale
- Logical, indicating whether to rescale the community names starting from 1. If FALSE, the community names will start from N+1, where N is the number of nodes.
Description
This function reconstructs the community tree of the community clustering analysis performed by the ‘cna’ function. It allows the user to explore different network community partitions.
Value
-
Returns a list object that includes the following components:
- modularity
- A numeric vector containing the modularity values.
- tree
- A numeric matrix containing in each row the community residue memberships corresponding to a modularity value. The rows are ordered according to the ‘modularity’ object.
- num.of.comms
- A numeric vector containing the number of communities per modularity value. The vector elements are ordered according to the ‘modularity’ object.
Details
The input of this function should be a ‘cna’ class object containing ‘network’ and ‘communities’ attributes.
This function reconstructs the community residue memberships for each modularity value. The purpose is to facilitate inspection of alternate community partitioning points, which in practice often corresponds to a value close to the maximum of the modularity, but not the maximum value itself.
Examples
# PDB server connection required - testing excluded ###-- Build a CNA object pdb <- read.pdb("4Q21")Note: Accessing on-line PDB filemodes <- nma(pdb)Building Hessian... Done in 0.063 seconds. Diagonalizing Hessian... Done in 0.277 seconds.cij <- dccm(modes)|======================================================================| 100%net <- cna(cij, cutoff.cij=0.2) ##-- Reconstruct the community membership vector for each clustering step. tree <- community.tree(net, rescale=TRUE) ## Plot modularity vs number of communities plot( tree$num.of.comms, tree$modularity ) ## Inspect the maximum modularity value partitioning max.mod.ind <- which.max(tree$modularity)
## Number of communities (k) at max modularity tree$num.of.comms[ max.mod.ind ][1] 9## Membership vector at this partition point tree$tree[max.mod.ind,][1] 1 1 1 1 1 2 2 2 3 3 3 3 3 3 4 4 4 4 4 4 4 4 4 4 4 4 4 4 4 4 4 4 2 2 2 2 2 [38] 2 2 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 5 5 5 5 5 5 5 5 5 5 5 5 6 5 [75] 5 5 2 2 7 3 3 3 8 8 8 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 9 9 [112] 7 3 3 8 8 8 8 8 8 8 8 8 8 7 7 7 7 7 7 7 7 7 7 7 7 7 7 7 7 7 7 8 8 8 4 4 4 [149] 4 8 8 8 8 8 9 9 9 9 9 9 9 9 9 9 9 9 9 9# Should be the same as that contained in the original CNA network object net$communities$membership == tree$tree[max.mod.ind,]1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 161 162 163 164 165 166 167 168 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE# Inspect a new membership partitioning (at k=7) memb.k7 <- tree$tree[ tree$num.of.comms == 7, ] ## Produce a new k=7 community network net.7 <- network.amendment(net, memb.k7) plot(net.7, pdb)Obtaining layout from PDB structure
#view.cna(net.7, trim.pdb(pdb, atom.select(pdb,"calpha")), launch=TRUE )