Integrating Protein Structural Dynamics and Evolutionary Analysis
29/09/14 14:53
I am the lead developer of bio3d, an R package for the exploratory analysis of structure and sequence data. Features of the package include the ability to read and write structure, sequence and dynamic trajectory data, perform atom summaries, atom selection, re-orientation, superposition, rigid core identification, clustering, distance matrix analysis, structure and sequence conservation analysis and principal component analysis. Bio3d takes advantage of the extensive graphical and statistical capabilities of the R environment and thus represents a useful framework for exploratory analysis of structural data. 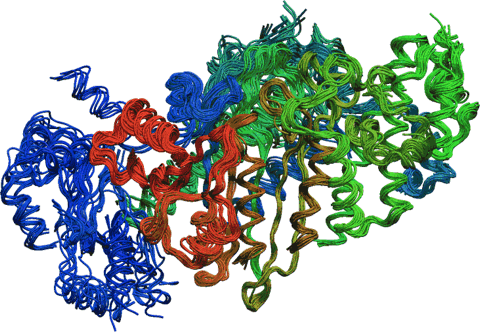
 The detailed comparison of homologous protein structures can be used to infer pathways for evolutionary adaptation and, at closer evolutionary distances, mechanisms for conformational change. Traditionally, such investigations have involved careful visual inspection combined with structural alignment methods. These procedures are both time consuming and labor intensive, and require expert insight into the systems studied. With the growing number of determined protein structures, the availability of automatic procedures for analyzing the differences and similarities between structures becomes increasingly desirable.
The detailed comparison of homologous protein structures can be used to infer pathways for evolutionary adaptation and, at closer evolutionary distances, mechanisms for conformational change. Traditionally, such investigations have involved careful visual inspection combined with structural alignment methods. These procedures are both time consuming and labor intensive, and require expert insight into the systems studied. With the growing number of determined protein structures, the availability of automatic procedures for analyzing the differences and similarities between structures becomes increasingly desirable.
Mouse over video for play/pause, full-screen options and description of analysis.
<< Back to Research Overview


Mouse over video for play/pause, full-screen options and description of analysis.
<< Back to Research Overview
