Conformational Selection & Allostery in Molecular Switches
29/09/14 14:53
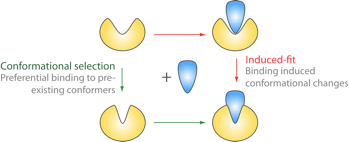
Monomeric G-proteins are a large superfamily of GTP-dependent cellular regulators. Composed of more than 6 functionally distinct families, these enzymes cycle between GTP- active and GDP-bound inactive conformations to regulate diverse functions from signal transduction and protein synthesis to cytoskeleletal remodeling. The critical activating conformational change common to all superfamily members has traditionally been described in terms of an induced-fit model, where the presence or absence of the γ-phosphate of GTP leads to an instantaneous switch in conformation.
Here we report evidence from atomistic molecular simulations that both Rho and Ras superfamily members harbor an intrinsic susceptibility to sample multiple conformational states regardless of the bound nucleotide. These results indicate that conformational changes in G-proteins may be best described by a population-shift mechanism rather than by the popular “loaded spring” or ligand induced-fit on/off switch model.
Furthermore these simulations together with in silico mutations provide evidence for a common preserved dynamic linkage between the nucleotide-binding site and the distal C-terminus critical for membrane interaction. The apparent allosteric coupling of these functional sites is likely to operate in a similar manor in all monomeric G-proteins.
<< Back to Research OverviewTags: NTPases, kinesin, ras, rho, md, amd, evolution