###
### Example of PCA on a collection of PKA structures
### and a large collection of transducin structure
###
### Authors Xin-Qiu Yao
### Lars Skjaerven
### Barry J Grant
###
require(bio3d); require(graphics);
pause <- function() {
cat("Press ENTER/RETURN/NEWLINE to continue.")
readLines(n=1)
invisible()
}
################################################
## #
## Basic PCA of related X-ray structures #
## (requires the 'muscle' program installed) #
## #
################################################
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Set temp dir to store PDB files
tmp.dir <- tempdir()
## Specify PDB identifiers
ids <- c("1cdk_A", "3agm_A", "1cmk_E",
"3dnd_A", "1q8w_A")
## Download PDBs
raw.files <- get.pdb(ids, path=tmp.dir)
pause()
Press ENTER/RETURN/NEWLINE to continue.
## Split PDBs by chain ID
files <- pdbsplit(raw.files, ids, path=tmp.dir)
|======================================================================| 100%
pause()
Press ENTER/RETURN/NEWLINE to continue.
## Sequence/structure alignment
pdbs <- pdbaln(files)
Reading PDB files:
/tmp/RtmpTDihxb/1cdk_A.pdb
/tmp/RtmpTDihxb/3agm_A.pdb
/tmp/RtmpTDihxb/1cmk_E.pdb
/tmp/RtmpTDihxb/3dnd_A.pdb
/tmp/RtmpTDihxb/1q8w_A.pdb
... PDB has ALT records, taking A only, rm.alt=TRUE
..
Extracting sequences
pdb/seq: 1 name: /tmp/RtmpTDihxb/1cdk_A.pdb
pdb/seq: 2 name: /tmp/RtmpTDihxb/3agm_A.pdb
pdb/seq: 3 name: /tmp/RtmpTDihxb/1cmk_E.pdb
pdb/seq: 4 name: /tmp/RtmpTDihxb/3dnd_A.pdb
PDB has ALT records, taking A only, rm.alt=TRUE
pdb/seq: 5 name: /tmp/RtmpTDihxb/1q8w_A.pdb
pause()
Press ENTER/RETURN/NEWLINE to continue.
## Find invariant core
core <- core.find(pdbs)
core size 335 of 336 vol = 23.654
core size 334 of 336 vol = 22.36
core size 333 of 336 vol = 21.426
core size 332 of 336 vol = 20.581
core size 331 of 336 vol = 19.576
core size 330 of 336 vol = 18.826
core size 329 of 336 vol = 18.16
core size 328 of 336 vol = 17.522
core size 327 of 336 vol = 16.914
core size 326 of 336 vol = 16.351
core size 325 of 336 vol = 15.8
core size 324 of 336 vol = 15.327
core size 323 of 336 vol = 14.79
core size 322 of 336 vol = 14.336
core size 321 of 336 vol = 14.02
core size 320 of 336 vol = 13.762
core size 319 of 336 vol = 13.48
core size 318 of 336 vol = 13.201
core size 317 of 336 vol = 12.918
core size 316 of 336 vol = 12.63
core size 315 of 336 vol = 12.395
core size 314 of 336 vol = 12.167
core size 313 of 336 vol = 12
core size 312 of 336 vol = 11.779
core size 311 of 336 vol = 11.613
core size 310 of 336 vol = 11.432
core size 309 of 336 vol = 11.277
core size 308 of 336 vol = 11.121
core size 307 of 336 vol = 10.891
core size 306 of 336 vol = 10.572
core size 305 of 336 vol = 10.333
core size 304 of 336 vol = 10.135
core size 303 of 336 vol = 9.98
core size 302 of 336 vol = 9.828
core size 301 of 336 vol = 9.665
core size 300 of 336 vol = 9.533
core size 299 of 336 vol = 9.275
core size 298 of 336 vol = 9.077
core size 297 of 336 vol = 8.906
core size 296 of 336 vol = 8.67
core size 295 of 336 vol = 8.459
core size 294 of 336 vol = 8.222
core size 293 of 336 vol = 8.075
core size 292 of 336 vol = 7.898
core size 291 of 336 vol = 7.781
core size 290 of 336 vol = 7.677
core size 289 of 336 vol = 7.5
core size 288 of 336 vol = 7.352
core size 287 of 336 vol = 7.245
core size 286 of 336 vol = 7.081
core size 285 of 336 vol = 6.91
core size 284 of 336 vol = 6.787
core size 283 of 336 vol = 6.665
core size 282 of 336 vol = 6.529
core size 281 of 336 vol = 6.379
core size 280 of 336 vol = 6.231
core size 279 of 336 vol = 6.142
core size 278 of 336 vol = 6.062
core size 277 of 336 vol = 5.997
core size 276 of 336 vol = 5.931
core size 275 of 336 vol = 5.836
core size 274 of 336 vol = 5.736
core size 273 of 336 vol = 5.652
core size 272 of 336 vol = 5.606
core size 271 of 336 vol = 5.543
core size 270 of 336 vol = 5.469
core size 269 of 336 vol = 5.388
core size 268 of 336 vol = 5.318
core size 267 of 336 vol = 5.23
core size 266 of 336 vol = 5.167
core size 265 of 336 vol = 5.099
core size 264 of 336 vol = 5.002
core size 263 of 336 vol = 4.944
core size 262 of 336 vol = 4.846
core size 261 of 336 vol = 4.755
core size 260 of 336 vol = 4.689
core size 259 of 336 vol = 4.626
core size 258 of 336 vol = 4.573
core size 257 of 336 vol = 4.522
core size 256 of 336 vol = 4.461
core size 255 of 336 vol = 4.398
core size 254 of 336 vol = 4.324
core size 253 of 336 vol = 4.268
core size 252 of 336 vol = 4.202
core size 251 of 336 vol = 4.113
core size 250 of 336 vol = 4.028
core size 249 of 336 vol = 3.953
core size 248 of 336 vol = 3.888
core size 247 of 336 vol = 3.8
core size 246 of 336 vol = 3.729
core size 245 of 336 vol = 3.63
core size 244 of 336 vol = 3.551
core size 243 of 336 vol = 3.482
core size 242 of 336 vol = 3.404
core size 241 of 336 vol = 3.323
core size 240 of 336 vol = 3.211
core size 239 of 336 vol = 3.145
core size 238 of 336 vol = 3.068
core size 237 of 336 vol = 2.967
core size 236 of 336 vol = 2.877
core size 235 of 336 vol = 2.795
core size 234 of 336 vol = 2.701
core size 233 of 336 vol = 2.614
core size 232 of 336 vol = 2.529
core size 231 of 336 vol = 2.448
core size 230 of 336 vol = 2.368
core size 229 of 336 vol = 2.297
core size 228 of 336 vol = 2.228
core size 227 of 336 vol = 2.142
core size 226 of 336 vol = 2.073
core size 225 of 336 vol = 2.008
core size 224 of 336 vol = 1.952
core size 223 of 336 vol = 1.896
core size 222 of 336 vol = 1.825
core size 221 of 336 vol = 1.759
core size 220 of 336 vol = 1.706
core size 219 of 336 vol = 1.659
core size 218 of 336 vol = 1.615
core size 217 of 336 vol = 1.559
core size 216 of 336 vol = 1.518
core size 215 of 336 vol = 1.476
core size 214 of 336 vol = 1.435
core size 213 of 336 vol = 1.401
core size 212 of 336 vol = 1.36
core size 211 of 336 vol = 1.328
core size 210 of 336 vol = 1.295
core size 209 of 336 vol = 1.259
core size 208 of 336 vol = 1.223
core size 207 of 336 vol = 1.181
core size 206 of 336 vol = 1.147
core size 205 of 336 vol = 1.116
core size 204 of 336 vol = 1.08
core size 203 of 336 vol = 1.051
core size 202 of 336 vol = 1.021
core size 201 of 336 vol = 0.994
core size 200 of 336 vol = 0.97
core size 199 of 336 vol = 0.947
core size 198 of 336 vol = 0.928
core size 197 of 336 vol = 0.907
core size 196 of 336 vol = 0.882
core size 195 of 336 vol = 0.853
core size 194 of 336 vol = 0.832
core size 193 of 336 vol = 0.808
core size 192 of 336 vol = 0.789
core size 191 of 336 vol = 0.763
core size 190 of 336 vol = 0.746
core size 189 of 336 vol = 0.728
core size 188 of 336 vol = 0.706
core size 187 of 336 vol = 0.686
core size 186 of 336 vol = 0.67
core size 185 of 336 vol = 0.655
core size 184 of 336 vol = 0.639
core size 183 of 336 vol = 0.622
core size 182 of 336 vol = 0.61
core size 181 of 336 vol = 0.595
core size 180 of 336 vol = 0.581
core size 179 of 336 vol = 0.565
core size 178 of 336 vol = 0.552
core size 177 of 336 vol = 0.536
core size 176 of 336 vol = 0.524
core size 175 of 336 vol = 0.511
core size 174 of 336 vol = 0.499
FINISHED: Min vol ( 0.5 ) reached
pause()
Press ENTER/RETURN/NEWLINE to continue.
## Fit structures to core region
xyz <- pdbfit(pdbs, inds=core$c1A.xyz)
## outpath="core_fit/", full.pdbs=T, het2atom=T)
pause()
Press ENTER/RETURN/NEWLINE to continue.
## Locate gap containing positions
gaps.pos <- gap.inspect(pdbs$xyz)
## Perform PCA on non-gap containing positions
pc.xray <- pca.xyz(xyz[,gaps.pos$f.inds])
pause()
Press ENTER/RETURN/NEWLINE to continue.
## Plot x-ray results
plot(pc.xray)
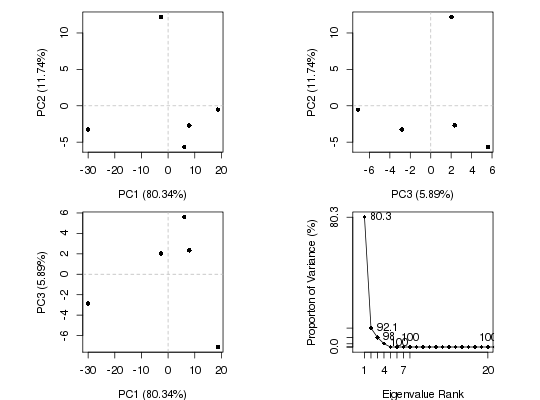
pause()
Press ENTER/RETURN/NEWLINE to continue.
#############################################
## #
## Larger transducin example #
## #
#############################################
data(transducin)
attach(transducin, warn.conflicts=FALSE)
## data 'transducin' contains objects
## - pdbs: aligned C-alpha coordinates for 53 transducin
## structures from the PDB
## - annotation: annotation of the 53 PDBs
## Note that this data can be generated from scratch by following the
## Comparative Structure Analysis with Bio3D Vignette available both
## on-line and from within the Bio3D package.
pdbs <- transducin$pdbs
annotation <- transducin$annotation
pause()
Press ENTER/RETURN/NEWLINE to continue.
## Inspect gaps
gaps.pos <- gap.inspect(pdbs$xyz)
## Previously fitted coordinates invariance core
xyz <- pdbs$xyz
## Do PCA
pc.xray <- pca.xyz(xyz[, gaps.pos$f.inds])
pause()
Press ENTER/RETURN/NEWLINE to continue.
## Plot overview
plot(pc.xray, col=annotation[, "color"])
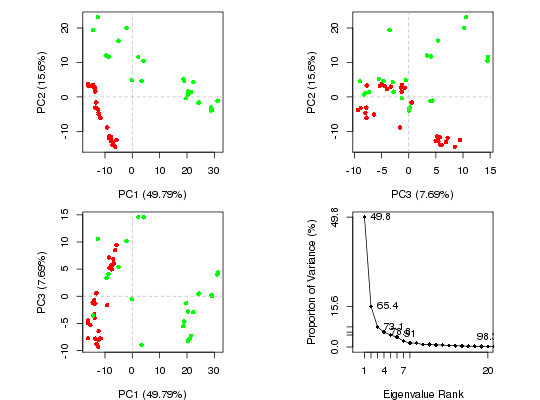
## Plot atom wise loadings
plot.bio3d(pc.xray$au[,1], ylab="PC1 (A)")
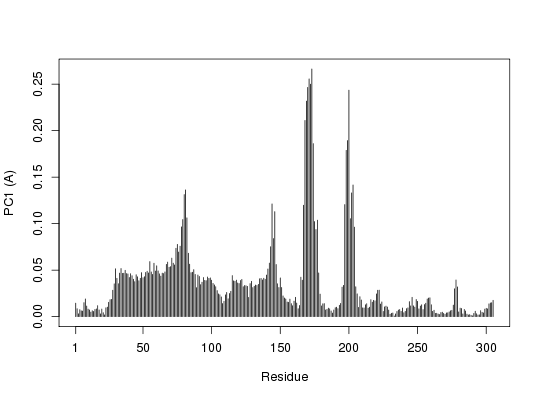
pause()
Press ENTER/RETURN/NEWLINE to continue.
unlink(tmp.dir)


