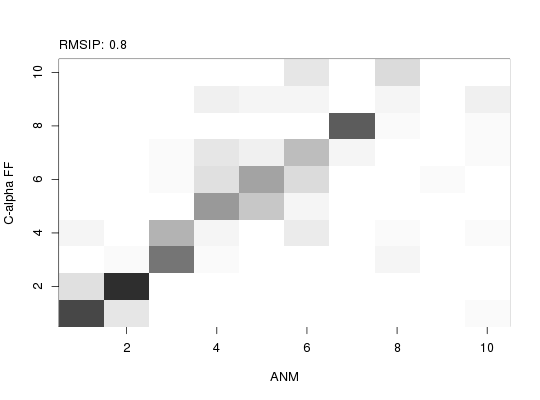###
### Examples from NMA Vignette
###
### Authors Lars Skjaerven
### Xin-Qiu Yao
### Barry J Grant
###
require(bio3d); require(graphics);
pause <- function() {
cat("Press ENTER/RETURN/NEWLINE to continue.")
readLines(n=1)
invisible()
}
#############################################
## #
## Basic usage #
## #
#############################################
### Read PDB and Calculate Normal Modes
pdb <- read.pdb("1hel")
Note: Accessing on-line PDB file
modes <- nma(pdb)
Building Hessian... Done in 0.046 seconds.
Diagonalizing Hessian... Done in 0.132 seconds.
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Print a summary
print(modes)
Call:
nma.pdb(pdb = pdb)
Class:
VibrationalModes (nma)
Number of modes:
387 (6 trivial)
Frequencies:
Mode 7: 0.018
Mode 8: 0.019
Mode 9: 0.024
Mode 10: 0.025
Mode 11: 0.028
Mode 12: 0.029
+ attr: modes, frequencies, force.constants, fluctuations,
U, L, xyz, mass, temp, triv.modes, natoms, call
pause()
Press ENTER/RETURN/NEWLINE to continue.
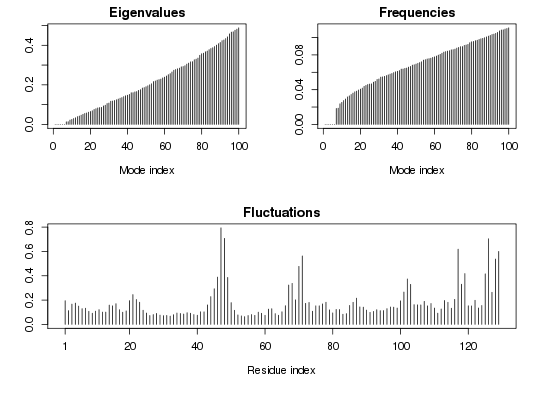
### Plot the nma object for a quick overview
plot(modes)
pause()
Press ENTER/RETURN/NEWLINE to continue.
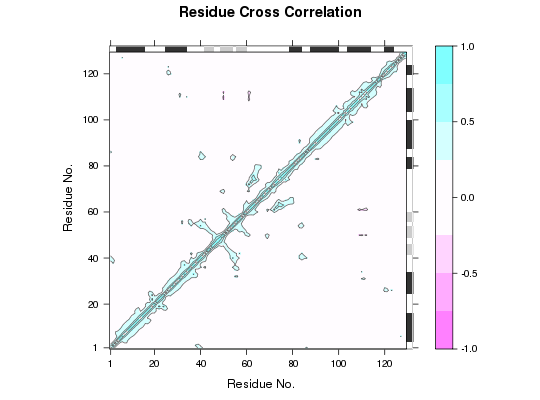
### Calculate cross-correlations
cm <- dccm(modes)
|======================================================================| 100%
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Plot correlation map
plot(cm, sse=pdb)
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Calculate modes with force field ANM
modes.anm <- nma(pdb, ff="anm")
Building Hessian... Done in 0.043 seconds.
Diagonalizing Hessian... Done in 0.134 seconds.
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Investigate modes similarity with RMSIP
r <- rmsip(modes, modes.anm)
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Plot RMSIP results
plot(r, xlab="ANM", ylab="C-alpha FF")
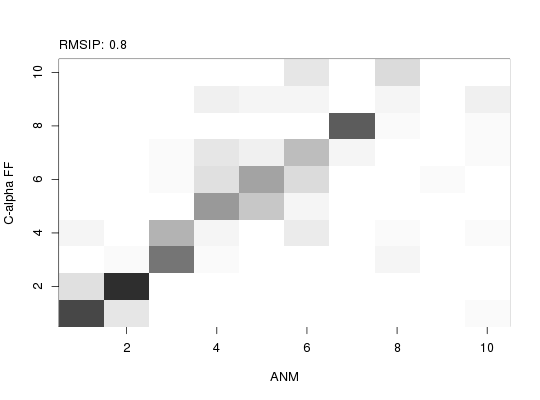
pause()
Press ENTER/RETURN/NEWLINE to continue.
################################################
## #
## Ensemble NMA #
## (requires the 'muscle' program installed) #
## #
################################################
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Set temp dir to store PDB files
tmp.dir <- tempdir()
### Download a set of DHFR structures
ids <- c("1rx2_A", "1rx4_A", "1rg7_A",
"3fyv_X", "3sgy_B")
### Download and split by chain ID
raw.files <- get.pdb(ids, path=tmp.dir)
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Split PDB files by chain ID
files <- pdbsplit( raw.files, ids, path=tmp.dir)
|======================================================================| 100%
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Align structures
pdbs <- pdbaln(files)
Reading PDB files:
/tmp/RtmpTDihxb/1rx2_A.pdb
/tmp/RtmpTDihxb/1rx4_A.pdb
/tmp/RtmpTDihxb/1rg7_A.pdb
/tmp/RtmpTDihxb/3fyv_X.pdb
/tmp/RtmpTDihxb/3sgy_B.pdb
PDB has ALT records, taking A only, rm.alt=TRUE
.....
Extracting sequences
pdb/seq: 1 name: /tmp/RtmpTDihxb/1rx2_A.pdb
PDB has ALT records, taking A only, rm.alt=TRUE
pdb/seq: 2 name: /tmp/RtmpTDihxb/1rx4_A.pdb
pdb/seq: 3 name: /tmp/RtmpTDihxb/1rg7_A.pdb
pdb/seq: 4 name: /tmp/RtmpTDihxb/3fyv_X.pdb
pdb/seq: 5 name: /tmp/RtmpTDihxb/3sgy_B.pdb
pause()
Press ENTER/RETURN/NEWLINE to continue.
### View sequence identity
summary( c(seqidentity(pdbs)) )
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.3590 0.3590 1.0000 0.6923 1.0000 1.0000
pause()
Press ENTER/RETURN/NEWLINE to continue.
### Calculate modes of aligned proteins
modes <- nma(pdbs)
Details of Scheduled Calculation:
... 5 input structures
... storing 462 eigenvectors for each structure
... dimension of x$U.subspace: ( 468x462x5 )
... coordinate superposition prior to NM calculation
... aligned eigenvectors (gap containing positions removed)
... estimated memory usage of final 'eNMA' object: 8.3 Mb
|======================================================================| 100%
pause()
Press ENTER/RETURN/NEWLINE to continue.
## Print a summary
print(modes)
Call:
nma.pdbs(pdbs = pdbs)
Class:
enma
Number of structures:
5
Attributes stored:
- Root mean square inner product (RMSIP)
- Aligned atomic fluctuations
- Aligned eigenvectors (gaps removed)
- Dimensions of x$U.subspace: 468x462x5
Coordinates were aligned prior to NMA calculations
+ attr: fluctuations, rmsip, U.subspace, L, full.nma, xyz,
call
pause()
Press ENTER/RETURN/NEWLINE to continue.
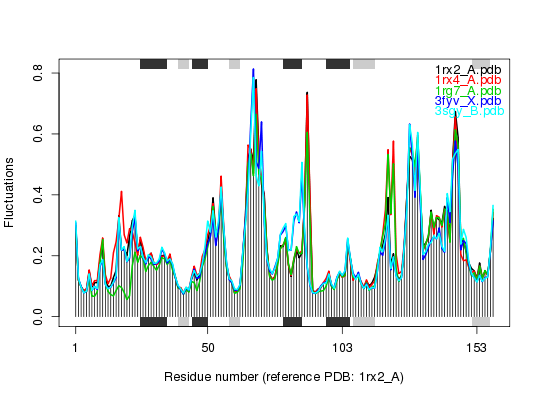
### Plot fluctuations
plot(modes, pdbs)
Extracting SSE from pdbs$sse attribute
pause()
Press ENTER/RETURN/NEWLINE to continue.
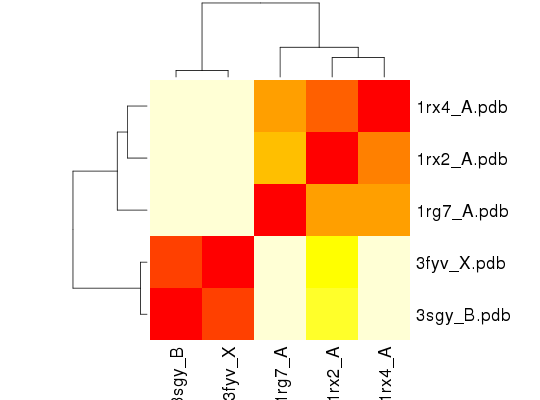
### Cluster Modes simiarlity
heatmap(1-modes$rmsip, labCol=ids)
unlink(tmp.dir)