Identification of Invariant Core Positions
Usage
core.find(...)"core.find"(pdbs, shortcut = FALSE, rm.island = FALSE, verbose = TRUE, stop.at = 15, stop.vol = 0.5, write.pdbs = FALSE, outpath="core_pruned", ncore = 1, nseg.scale = 1, ...)"core.find"(xyz, ...)"core.find"(pdb, verbose=TRUE, ...)
Arguments
- pdbs
- a numeric matrix of aligned C-alpha xyz Cartesian
coordinates. For example an alignment data structure obtained with
read.fasta.pdborpdbaln. - shortcut
- if TRUE, remove more than one position at a time.
- rm.island
- remove isolated fragments of less than three residues.
- verbose
- logical, if TRUE a “core\_pruned” directory containing ‘core structures’ for each iteraction is written to the current directory.
- stop.at
- minimal core size at which iterations should be stopped.
- stop.vol
- minimal core volume at which iterations should be stopped.
- write.pdbs
- logical, if TRUE core coordinate files, containing
only core positions for each iteration, are written to a location
specified by
outpath. - outpath
- character string specifying the output directory when
write.pdbsis TRUE. - ncore
- number of CPU cores used to do the calculation.
ncore>1requires package ‘parallel’ installed. - nseg.scale
- split input data into specified number of segments
prior to running multiple core calculation. See
fit.xyz. - xyz
- a numeric matrix of xyz Cartesian coordinates,
e.g. obtained from
read.dcdorread.ncdf. - pdb
- an object of type
pdbas obtained from functionread.pdbwith multiple frames (>=4) stored in itsxyzcomponent. Note that the function will attempt to identify C-alpha and phosphate atoms (for protein and nucleic acids, respectively) in which the calculation should be based. - ...
- arguments passed to and from functions.
Description
Perform iterated rounds of structural superposition to identify the most invariant region in an aligned set of protein structures.
Details
This function attempts to iteratively refine an initial structural superposition determined from a multiple alignment. This involves iterated rounds of superposition, where at each round the position(s) displaying the largest differences is(are) excluded from the dataset. The spatial variation at each aligned position is determined from the eigenvalues of their Cartesian coordinates (i.e. the variance of the distribution along its three principal directions). Inspired by the work of Gerstein et al. (1991, 1995), an ellipsoid of variance is determined from the eigenvalues, and its volume is taken as a measure of structural variation at a given position.
Optional “core PDB files” containing core positions, upon which
superposition is based, can be written to a location specified by
outpath by setting write.pdbs=TRUE. These files are
useful for examining the core filtering process by visualising them in a
graphics program.
Value
-
Returns a list of class
- volume
- total core volume at each fitting iteration/round.
- length
- core length at each round.
- resno
- residue number of core residues at each round (taken from the first aligned structure) or, alternatively, the numeric index of core residues at each round.
- step.inds
- atom indices of core atoms at each round.
- atom
- atom indices of core positions in the last round.
- xyz
- xyz indices of core positions in the last round.
- c1A.atom
- atom indices of core positions with a total volume under 1 Angstrom\^3.
- c1A.xyz
- xyz indices of core positions with a total volume under 1 Angstrom\^3.
- c1A.resno
- residue numbers of core positions with a total volume under 1 Angstrom\^3.
- c0.5A.atom
- atom indices of core positions with a total volume under 0.5 Angstrom\^3.
- c0.5A.xyz
- xyz indices of core positions with a total volume under 0.5 Angstrom\^3.
- c0.5A.resno
- residue numbers of core positions with a total volume under 0.5 Angstrom\^3.
"core" with the following components:
References
Grant, B.J. et al. (2006) Bioinformatics 22, 2695--2696.
Gerstein and Altman (1995) J. Mol. Biol. 251, 161--175.
Gerstein and Chothia (1991) J. Mol. Biol. 220, 133--149.
Note
The relevance of the ‘core positions’ identified by this procedure is dependent upon the number of input structures and their diversity.
Examples
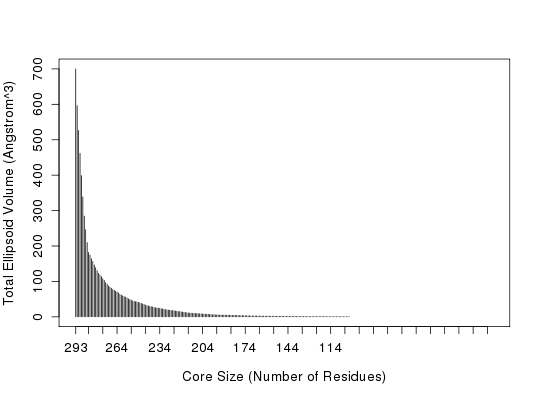
##-- Generate a small kinesin alignment and read corresponding structures pdbfiles <- get.pdb(c("1bg2","2ncd","1i6i","1i5s"), URLonly=TRUE) pdbs <- pdbaln(pdbfiles)Reading PDB files: http://www.rcsb.org/pdb/files/1bg2.pdb http://www.rcsb.org/pdb/files/2ncd.pdb http://www.rcsb.org/pdb/files/1i6i.pdb http://www.rcsb.org/pdb/files/1i5s.pdb .. PDB has ALT records, taking A only, rm.alt=TRUE . PDB has ALT records, taking A only, rm.alt=TRUE . Extracting sequences pdb/seq: 1 name: http://www.rcsb.org/pdb/files/1bg2.pdb pdb/seq: 2 name: http://www.rcsb.org/pdb/files/2ncd.pdb pdb/seq: 3 name: http://www.rcsb.org/pdb/files/1i6i.pdb PDB has ALT records, taking A only, rm.alt=TRUE pdb/seq: 4 name: http://www.rcsb.org/pdb/files/1i5s.pdb PDB has ALT records, taking A only, rm.alt=TRUE##-- Find 'core' positions core <- core.find(pdbs)core size 293 of 294 vol = 699.925 core size 292 of 294 vol = 596.377 core size 291 of 294 vol = 526.367 core size 290 of 294 vol = 461.809 core size 289 of 294 vol = 398.743 core size 288 of 294 vol = 338.511 core size 287 of 294 vol = 284.55 core size 286 of 294 vol = 245.99 core size 285 of 294 vol = 209.855 core size 284 of 294 vol = 181.912 core size 283 of 294 vol = 174.392 core size 282 of 294 vol = 164.476 core size 281 of 294 vol = 156.414 core size 280 of 294 vol = 146.519 core size 279 of 294 vol = 138.917 core size 278 of 294 vol = 130.52 core size 277 of 294 vol = 123.34 core size 276 of 294 vol = 118.842 core size 275 of 294 vol = 113.597 core size 274 of 294 vol = 107.865 core size 273 of 294 vol = 103.215 core size 272 of 294 vol = 97.239 core size 271 of 294 vol = 92.829 core size 270 of 294 vol = 87.717 core size 269 of 294 vol = 84.086 core size 268 of 294 vol = 80.571 core size 267 of 294 vol = 77.329 core size 266 of 294 vol = 75.464 core size 265 of 294 vol = 72.27 core size 264 of 294 vol = 70.298 core size 263 of 294 vol = 67.809 core size 262 of 294 vol = 64.211 core size 261 of 294 vol = 61.395 core size 260 of 294 vol = 59.047 core size 259 of 294 vol = 57.234 core size 258 of 294 vol = 55.442 core size 257 of 294 vol = 53.374 core size 256 of 294 vol = 50.983 core size 255 of 294 vol = 48.827 core size 254 of 294 vol = 47.141 core size 253 of 294 vol = 45.141 core size 252 of 294 vol = 43.701 core size 251 of 294 vol = 43.009 core size 250 of 294 vol = 42.575 core size 249 of 294 vol = 40.857 core size 248 of 294 vol = 39.521 core size 247 of 294 vol = 38.055 core size 246 of 294 vol = 36.383 core size 245 of 294 vol = 34.777 core size 244 of 294 vol = 33.234 core size 243 of 294 vol = 31.942 core size 242 of 294 vol = 30.77 core size 241 of 294 vol = 29.364 core size 240 of 294 vol = 28.579 core size 239 of 294 vol = 27.857 core size 238 of 294 vol = 26.662 core size 237 of 294 vol = 25.791 core size 236 of 294 vol = 25.126 core size 235 of 294 vol = 24.758 core size 234 of 294 vol = 24.072 core size 233 of 294 vol = 22.984 core size 232 of 294 vol = 22.483 core size 231 of 294 vol = 21.414 core size 230 of 294 vol = 20.533 core size 229 of 294 vol = 19.881 core size 228 of 294 vol = 19.328 core size 227 of 294 vol = 18.594 core size 226 of 294 vol = 17.918 core size 225 of 294 vol = 17.824 core size 224 of 294 vol = 17.519 core size 223 of 294 vol = 16.291 core size 222 of 294 vol = 15.704 core size 221 of 294 vol = 15.036 core size 220 of 294 vol = 14.852 core size 219 of 294 vol = 14.298 core size 218 of 294 vol = 13.536 core size 217 of 294 vol = 12.802 core size 216 of 294 vol = 12.249 core size 215 of 294 vol = 11.663 core size 214 of 294 vol = 11.121 core size 213 of 294 vol = 10.52 core size 212 of 294 vol = 10.379 core size 211 of 294 vol = 10.062 core size 210 of 294 vol = 9.795 core size 209 of 294 vol = 9.539 core size 208 of 294 vol = 9.176 core size 207 of 294 vol = 9.299 core size 206 of 294 vol = 8.77 core size 205 of 294 vol = 8.437 core size 204 of 294 vol = 8.153 core size 203 of 294 vol = 7.771 core size 202 of 294 vol = 7.517 core size 201 of 294 vol = 7.312 core size 200 of 294 vol = 7.091 core size 199 of 294 vol = 6.829 core size 198 of 294 vol = 6.458 core size 197 of 294 vol = 6.261 core size 196 of 294 vol = 6.101 core size 195 of 294 vol = 5.734 core size 194 of 294 vol = 5.563 core size 193 of 294 vol = 5.372 core size 192 of 294 vol = 5.228 core size 191 of 294 vol = 5.153 core size 190 of 294 vol = 5.03 core size 189 of 294 vol = 4.917 core size 188 of 294 vol = 4.85 core size 187 of 294 vol = 4.744 core size 186 of 294 vol = 4.613 core size 185 of 294 vol = 4.478 core size 184 of 294 vol = 4.39 core size 183 of 294 vol = 4.275 core size 182 of 294 vol = 4.149 core size 181 of 294 vol = 4.034 core size 180 of 294 vol = 3.944 core size 179 of 294 vol = 3.876 core size 178 of 294 vol = 3.753 core size 177 of 294 vol = 3.753 core size 176 of 294 vol = 3.621 core size 175 of 294 vol = 3.457 core size 174 of 294 vol = 3.349 core size 173 of 294 vol = 3.119 core size 172 of 294 vol = 2.994 core size 171 of 294 vol = 2.875 core size 170 of 294 vol = 2.736 core size 169 of 294 vol = 2.681 core size 168 of 294 vol = 2.591 core size 167 of 294 vol = 2.535 core size 166 of 294 vol = 2.475 core size 165 of 294 vol = 2.43 core size 164 of 294 vol = 2.383 core size 163 of 294 vol = 2.351 core size 162 of 294 vol = 2.193 core size 161 of 294 vol = 2.134 core size 160 of 294 vol = 2.086 core size 159 of 294 vol = 2.042 core size 158 of 294 vol = 1.952 core size 157 of 294 vol = 1.908 core size 156 of 294 vol = 1.854 core size 155 of 294 vol = 1.814 core size 154 of 294 vol = 1.761 core size 153 of 294 vol = 1.729 core size 152 of 294 vol = 1.672 core size 151 of 294 vol = 1.631 core size 150 of 294 vol = 1.589 core size 149 of 294 vol = 1.537 core size 148 of 294 vol = 1.509 core size 147 of 294 vol = 1.496 core size 146 of 294 vol = 1.474 core size 145 of 294 vol = 1.449 core size 144 of 294 vol = 1.365 core size 143 of 294 vol = 1.33 core size 142 of 294 vol = 1.303 core size 141 of 294 vol = 1.268 core size 140 of 294 vol = 1.268 core size 139 of 294 vol = 1.24 core size 138 of 294 vol = 1.231 core size 137 of 294 vol = 1.214 core size 136 of 294 vol = 1.119 core size 135 of 294 vol = 1.112 core size 134 of 294 vol = 1.102 core size 133 of 294 vol = 1.078 core size 132 of 294 vol = 1.043 core size 131 of 294 vol = 1.018 core size 130 of 294 vol = 0.969 core size 129 of 294 vol = 0.938 core size 128 of 294 vol = 0.93 core size 127 of 294 vol = 0.903 core size 126 of 294 vol = 0.882 core size 125 of 294 vol = 0.873 core size 124 of 294 vol = 0.86 core size 123 of 294 vol = 0.851 core size 122 of 294 vol = 0.833 core size 121 of 294 vol = 0.824 core size 120 of 294 vol = 0.803 core size 119 of 294 vol = 0.78 core size 118 of 294 vol = 0.756 core size 117 of 294 vol = 0.763 core size 116 of 294 vol = 0.712 core size 115 of 294 vol = 0.704 core size 114 of 294 vol = 0.697 core size 113 of 294 vol = 0.665 core size 112 of 294 vol = 0.656 core size 111 of 294 vol = 0.64 core size 110 of 294 vol = 0.632 core size 109 of 294 vol = 0.608 core size 108 of 294 vol = 0.594 core size 107 of 294 vol = 0.584 core size 106 of 294 vol = 0.528 core size 105 of 294 vol = 0.526 core size 104 of 294 vol = 0.516 core size 103 of 294 vol = 0.534 core size 102 of 294 vol = 0.517 core size 101 of 294 vol = 0.485 FINISHED: Min vol ( 0.5 ) reachedplot(core) ##-- Fit on these relatively invarient subset of positions #core.inds <- print(core, vol=1) core.inds <- print(core, vol=0.5)
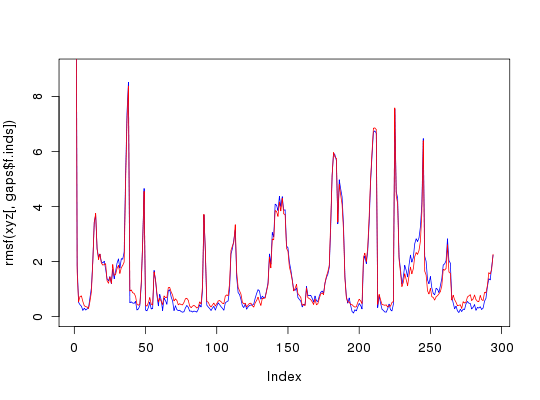
# 102 positions (cumulative volume <= 0.5 Angstrom^3) start end length 1 9 16 8 2 34 34 1 3 36 36 1 4 51 53 3 5 57 59 3 6 61 62 2 7 67 67 1 8 70 70 1 9 72 72 1 10 74 74 1 11 78 78 1 12 80 95 16 13 97 97 1 14 104 104 1 15 106 118 13 16 129 129 1 17 132 134 3 18 136 137 2 19 168 168 1 20 172 173 2 21 179 179 1 22 181 181 1 23 183 183 1 24 205 210 6 25 212 213 2 26 225 225 1 27 228 236 9 28 284 284 1 29 294 294 1 30 296 303 8 31 306 306 1 32 308 309 2 33 311 313 3 34 315 316 2xyz <- pdbfit(pdbs, core.inds, outpath="corefit_structures")PDB has ALT records, taking A only, rm.alt=TRUE PDB has ALT records, taking A only, rm.alt=TRUE##-- Compare to fitting on all equivalent positions xyz2 <- pdbfit(pdbs) ## Note that overall RMSD will be higher but RMSF will ## be lower in core regions, which may equate to a ## 'better fit' for certain applications gaps <- gap.inspect(pdbs$xyz) rmsd(xyz[,gaps$f.inds])Warning message: No indices provided, using the 294 non NA positions[,1] [,2] [,3] [,4] [1,] 0.000 3.583 3.390 3.116 [2,] 3.583 0.000 3.787 3.599 [3,] 3.390 3.787 0.000 2.351 [4,] 3.116 3.599 2.351 0.000rmsd(xyz2[,gaps$f.inds])Warning message: No indices provided, using the 294 non NA positions[,1] [,2] [,3] [,4] [1,] 0.000 3.570 3.297 3.089 [2,] 3.570 0.000 3.696 3.579 [3,] 3.297 3.696 0.000 2.222 [4,] 3.089 3.579 2.222 0.000plot(rmsf(xyz[,gaps$f.inds]), typ="l", col="blue", ylim=c(0,9))points(rmsf(xyz2[,gaps$f.inds]), typ="l", col="red")
##-- Run core.find() on a multimodel PDB file pdb <- read.pdb('1d1d', multi=TRUE)Note: Accessing on-line PDB filecore <- core.find(pdb).. 220 atom(s) from 'string' selection .. 220 atom(s) in final combined selection .. 0 atom(s) from 'string' selection .. 0 atom(s) from 'elety' selection .. 0 atom(s) in final combined selection Union of selects * Selected a total of: 220 atoms * core size 219 of 220 vol = 80028.92 core size 218 of 220 vol = 78588.22 core size 217 of 220 vol = 76410.13 core size 216 of 220 vol = 74091.53 core size 215 of 220 vol = 72626.57 core size 214 of 220 vol = 70791.32 core size 213 of 220 vol = 69224.37 core size 212 of 220 vol = 67861.81 core size 211 of 220 vol = 66479.53 core size 210 of 220 vol = 64913.15 core size 209 of 220 vol = 63452.38 core size 208 of 220 vol = 62054.96 core size 207 of 220 vol = 60965.03 core size 206 of 220 vol = 59532.15 core size 205 of 220 vol = 58123.13 core size 204 of 220 vol = 56913.48 core size 203 of 220 vol = 56013.92 core size 202 of 220 vol = 54824.84 core size 201 of 220 vol = 53692.84 core size 200 of 220 vol = 51976.8 core size 199 of 220 vol = 50319.93 core size 198 of 220 vol = 49132.73 core size 197 of 220 vol = 48361.34 core size 196 of 220 vol = 47276.21 core size 195 of 220 vol = 46269.85 core size 194 of 220 vol = 45136.55 core size 193 of 220 vol = 44072.71 core size 192 of 220 vol = 43323.08 core size 191 of 220 vol = 42364.85 core size 190 of 220 vol = 41325.58 core size 189 of 220 vol = 40658.42 core size 188 of 220 vol = 39536.91 core size 187 of 220 vol = 38598.77 core size 186 of 220 vol = 37659.78 core size 185 of 220 vol = 36891.51 core size 184 of 220 vol = 36240.27 core size 183 of 220 vol = 35560.02 core size 182 of 220 vol = 34684.24 core size 181 of 220 vol = 33889.36 core size 180 of 220 vol = 33212.81 core size 179 of 220 vol = 32233.6 core size 178 of 220 vol = 31223.56 core size 177 of 220 vol = 30556.8 core size 176 of 220 vol = 29908.46 core size 175 of 220 vol = 29253.3 core size 174 of 220 vol = 28070.19 core size 173 of 220 vol = 27319.49 core size 172 of 220 vol = 26595.86 core size 171 of 220 vol = 25931.99 core size 170 of 220 vol = 25254.61 core size 169 of 220 vol = 24538.45 core size 168 of 220 vol = 24041.35 core size 167 of 220 vol = 23064.07 core size 166 of 220 vol = 22528.61 core size 165 of 220 vol = 22040.96 core size 164 of 220 vol = 21487.35 core size 163 of 220 vol = 20773.03 core size 162 of 220 vol = 20172.09 core size 161 of 220 vol = 19541.43 core size 160 of 220 vol = 19063.78 core size 159 of 220 vol = 18534.67 core size 158 of 220 vol = 18075.66 core size 157 of 220 vol = 17488.76 core size 156 of 220 vol = 17003.33 core size 155 of 220 vol = 16423.39 core size 154 of 220 vol = 15648.52 core size 153 of 220 vol = 14945.91 core size 152 of 220 vol = 13975.75 core size 151 of 220 vol = 13291.79 core size 150 of 220 vol = 12455.54 core size 149 of 220 vol = 11440.64 core size 148 of 220 vol = 10332.09 core size 147 of 220 vol = 9491.928 core size 146 of 220 vol = 8548.282 core size 145 of 220 vol = 7373.966 core size 144 of 220 vol = 6194.034 core size 143 of 220 vol = 4856.297 core size 142 of 220 vol = 3238.08 core size 141 of 220 vol = 1915.693 core size 140 of 220 vol = 1059.597 core size 139 of 220 vol = 599.305 core size 138 of 220 vol = 383.745 core size 137 of 220 vol = 256.211 core size 136 of 220 vol = 158.747 core size 135 of 220 vol = 107.812 core size 134 of 220 vol = 88.012 core size 133 of 220 vol = 69.301 core size 132 of 220 vol = 55.639 core size 131 of 220 vol = 44.668 core size 130 of 220 vol = 33.939 core size 129 of 220 vol = 26.558 core size 128 of 220 vol = 24.122 core size 127 of 220 vol = 22.316 core size 126 of 220 vol = 21.486 core size 125 of 220 vol = 20.677 core size 124 of 220 vol = 19.789 core size 123 of 220 vol = 18.956 core size 122 of 220 vol = 18.03 core size 121 of 220 vol = 17.241 core size 120 of 220 vol = 16.465 core size 119 of 220 vol = 15.771 core size 118 of 220 vol = 15.084 core size 117 of 220 vol = 14.441 core size 116 of 220 vol = 13.958 core size 115 of 220 vol = 13.364 core size 114 of 220 vol = 12.795 core size 113 of 220 vol = 12.228 core size 112 of 220 vol = 11.703 core size 111 of 220 vol = 11.166 core size 110 of 220 vol = 10.723 core size 109 of 220 vol = 10.251 core size 108 of 220 vol = 9.788 core size 107 of 220 vol = 9.355 core size 106 of 220 vol = 8.913 core size 105 of 220 vol = 8.475 core size 104 of 220 vol = 8.113 core size 103 of 220 vol = 7.737 core size 102 of 220 vol = 7.436 core size 101 of 220 vol = 7.024 core size 100 of 220 vol = 6.712 core size 99 of 220 vol = 6.403 core size 98 of 220 vol = 6.11 core size 97 of 220 vol = 5.777 core size 96 of 220 vol = 5.467 core size 95 of 220 vol = 5.087 core size 94 of 220 vol = 4.73 core size 93 of 220 vol = 4.44 core size 92 of 220 vol = 4.139 core size 91 of 220 vol = 3.814 core size 90 of 220 vol = 3.544 core size 89 of 220 vol = 3.336 core size 88 of 220 vol = 3.17 core size 87 of 220 vol = 2.99 core size 86 of 220 vol = 2.81 core size 85 of 220 vol = 2.613 core size 84 of 220 vol = 2.436 core size 83 of 220 vol = 2.269 core size 82 of 220 vol = 2.124 core size 81 of 220 vol = 1.999 core size 80 of 220 vol = 1.899 core size 79 of 220 vol = 1.8 core size 78 of 220 vol = 1.726 core size 77 of 220 vol = 1.662 core size 76 of 220 vol = 1.578 core size 75 of 220 vol = 1.511 core size 74 of 220 vol = 1.451 core size 73 of 220 vol = 1.394 core size 72 of 220 vol = 1.337 core size 71 of 220 vol = 1.289 core size 70 of 220 vol = 1.239 core size 69 of 220 vol = 1.186 core size 68 of 220 vol = 1.137 core size 67 of 220 vol = 1.094 core size 66 of 220 vol = 1.044 core size 65 of 220 vol = 0.995 core size 64 of 220 vol = 0.947 core size 63 of 220 vol = 0.9 core size 62 of 220 vol = 0.858 core size 61 of 220 vol = 0.81 core size 60 of 220 vol = 0.767 core size 59 of 220 vol = 0.733 core size 58 of 220 vol = 0.697 core size 57 of 220 vol = 0.659 core size 56 of 220 vol = 0.628 core size 55 of 220 vol = 0.601 core size 54 of 220 vol = 0.563 core size 53 of 220 vol = 0.527 core size 52 of 220 vol = 0.491 FINISHED: Min vol ( 0.5 ) reached##-- Run core.find() on a trajectory trtfile <- system.file("examples/hivp.dcd", package="bio3d") trj <- read.dcd(trtfile)NATOM = 198 NFRAME= 117 ISTART= 0 last = 117 nstep = 117 nfile = 117 NSAVE = 1 NDEGF = 0 version 24 Reading (x100) |======================================================================| 100%## Read the starting PDB file to determine atom correspondence pdbfile <- system.file("examples/hivp.pdb", package="bio3d") pdb <- read.pdb(pdbfile) ## select calpha coords from a manageable number of frames ca.ind <- atom.select(pdb, "calpha")$xyz frames <- seq(1, nrow(trj), by=10) core <- core.find( trj[frames, ca.ind], write.pdbs=TRUE )core size 197 of 198 vol = 379.113 core size 196 of 198 vol = 359.625 core size 195 of 198 vol = 342.013 core size 194 of 198 vol = 327.636 core size 193 of 198 vol = 314.284 core size 192 of 198 vol = 302.926 core size 191 of 198 vol = 291.664 core size 190 of 198 vol = 280.909 core size 189 of 198 vol = 270.866 core size 188 of 198 vol = 261.247 core size 187 of 198 vol = 252.37 core size 186 of 198 vol = 243.948 core size 185 of 198 vol = 235.569 core size 184 of 198 vol = 227.501 core size 183 of 198 vol = 220.684 core size 182 of 198 vol = 213.842 core size 181 of 198 vol = 206.979 core size 180 of 198 vol = 200.46 core size 179 of 198 vol = 194.523 core size 178 of 198 vol = 189.055 core size 177 of 198 vol = 183.923 core size 176 of 198 vol = 178.328 core size 175 of 198 vol = 173.85 core size 174 of 198 vol = 169.616 core size 173 of 198 vol = 164.985 core size 172 of 198 vol = 160.923 core size 171 of 198 vol = 156.433 core size 170 of 198 vol = 152.544 core size 169 of 198 vol = 148.882 core size 168 of 198 vol = 144.709 core size 167 of 198 vol = 141.148 core size 166 of 198 vol = 137.941 core size 165 of 198 vol = 134.225 core size 164 of 198 vol = 130.687 core size 163 of 198 vol = 127.211 core size 162 of 198 vol = 124.155 core size 161 of 198 vol = 121.213 core size 160 of 198 vol = 118.4 core size 159 of 198 vol = 116.104 core size 158 of 198 vol = 113.746 core size 157 of 198 vol = 111.249 core size 156 of 198 vol = 108.6 core size 155 of 198 vol = 106.528 core size 154 of 198 vol = 104.057 core size 153 of 198 vol = 102.063 core size 152 of 198 vol = 100.064 core size 151 of 198 vol = 97.77 core size 150 of 198 vol = 95.843 core size 149 of 198 vol = 94.004 core size 148 of 198 vol = 92.017 core size 147 of 198 vol = 90.19 core size 146 of 198 vol = 88.477 core size 145 of 198 vol = 86.675 core size 144 of 198 vol = 84.823 core size 143 of 198 vol = 83.159 core size 142 of 198 vol = 81.642 core size 141 of 198 vol = 80.064 core size 140 of 198 vol = 78.276 core size 139 of 198 vol = 76.764 core size 138 of 198 vol = 75.33 core size 137 of 198 vol = 73.963 core size 136 of 198 vol = 72.652 core size 135 of 198 vol = 71.334 core size 134 of 198 vol = 69.968 core size 133 of 198 vol = 68.612 core size 132 of 198 vol = 66.996 core size 131 of 198 vol = 65.66 core size 130 of 198 vol = 64.271 core size 129 of 198 vol = 62.969 core size 128 of 198 vol = 61.783 core size 127 of 198 vol = 60.592 core size 126 of 198 vol = 59.539 core size 125 of 198 vol = 58.411 core size 124 of 198 vol = 57.264 core size 123 of 198 vol = 56.185 core size 122 of 198 vol = 55.117 core size 121 of 198 vol = 54.052 core size 120 of 198 vol = 52.987 core size 119 of 198 vol = 51.928 core size 118 of 198 vol = 50.868 core size 117 of 198 vol = 49.942 core size 116 of 198 vol = 48.97 core size 115 of 198 vol = 47.903 core size 114 of 198 vol = 46.909 core size 113 of 198 vol = 45.909 core size 112 of 198 vol = 44.944 core size 111 of 198 vol = 43.991 core size 110 of 198 vol = 43.068 core size 109 of 198 vol = 42.121 core size 108 of 198 vol = 41.071 core size 107 of 198 vol = 40.146 core size 106 of 198 vol = 39.336 core size 105 of 198 vol = 38.507 core size 104 of 198 vol = 37.752 core size 103 of 198 vol = 36.972 core size 102 of 198 vol = 36.345 core size 101 of 198 vol = 35.598 core size 100 of 198 vol = 34.916 core size 99 of 198 vol = 34.26 core size 98 of 198 vol = 33.578 core size 97 of 198 vol = 32.882 core size 96 of 198 vol = 32.107 core size 95 of 198 vol = 31.456 core size 94 of 198 vol = 30.663 core size 93 of 198 vol = 30.01 core size 92 of 198 vol = 29.393 core size 91 of 198 vol = 28.798 core size 90 of 198 vol = 28.159 core size 89 of 198 vol = 27.491 core size 88 of 198 vol = 26.833 core size 87 of 198 vol = 26.209 core size 86 of 198 vol = 25.625 core size 85 of 198 vol = 25.025 core size 84 of 198 vol = 24.448 core size 83 of 198 vol = 23.778 core size 82 of 198 vol = 23.211 core size 81 of 198 vol = 22.589 core size 80 of 198 vol = 22.006 core size 79 of 198 vol = 21.436 core size 78 of 198 vol = 20.869 core size 77 of 198 vol = 20.402 core size 76 of 198 vol = 19.866 core size 75 of 198 vol = 19.282 core size 74 of 198 vol = 18.73 core size 73 of 198 vol = 18.261 core size 72 of 198 vol = 17.768 core size 71 of 198 vol = 17.366 core size 70 of 198 vol = 16.903 core size 69 of 198 vol = 16.356 core size 68 of 198 vol = 15.898 core size 67 of 198 vol = 15.478 core size 66 of 198 vol = 15.068 core size 65 of 198 vol = 14.652 core size 64 of 198 vol = 14.184 core size 63 of 198 vol = 13.798 core size 62 of 198 vol = 13.391 core size 61 of 198 vol = 12.968 core size 60 of 198 vol = 12.587 core size 59 of 198 vol = 12.139 core size 58 of 198 vol = 11.711 core size 57 of 198 vol = 11.329 core size 56 of 198 vol = 10.93 core size 55 of 198 vol = 10.555 core size 54 of 198 vol = 10.143 core size 53 of 198 vol = 9.74 core size 52 of 198 vol = 9.289 core size 51 of 198 vol = 8.917 core size 50 of 198 vol = 8.587 core size 49 of 198 vol = 8.248 core size 48 of 198 vol = 7.925 core size 47 of 198 vol = 7.618 core size 46 of 198 vol = 7.365 core size 45 of 198 vol = 7.027 core size 44 of 198 vol = 6.723 core size 43 of 198 vol = 6.461 core size 42 of 198 vol = 6.186 core size 41 of 198 vol = 5.969 core size 40 of 198 vol = 5.771 core size 39 of 198 vol = 5.489 core size 38 of 198 vol = 5.21 core size 37 of 198 vol = 4.932 core size 36 of 198 vol = 4.676 core size 35 of 198 vol = 4.376 core size 34 of 198 vol = 4.145 core size 33 of 198 vol = 3.898 core size 32 of 198 vol = 3.707 core size 31 of 198 vol = 3.458 core size 30 of 198 vol = 3.229 core size 29 of 198 vol = 3.02 core size 28 of 198 vol = 2.834 core size 27 of 198 vol = 2.638 core size 26 of 198 vol = 2.448 core size 25 of 198 vol = 2.269 core size 24 of 198 vol = 2.06 core size 23 of 198 vol = 1.893 core size 22 of 198 vol = 1.698 core size 21 of 198 vol = 1.517 core size 20 of 198 vol = 1.341 core size 19 of 198 vol = 1.218 core size 18 of 198 vol = 1.072 core size 17 of 198 vol = 0.954 core size 16 of 198 vol = 0.822 core size 15 of 198 vol = 0.744## have a look at the various cores "vmd -m core_pruned/*.pdb" ## Lets use a 6A^3 core cutoff inds <- print(core, vol=6)# 42 positions (cumulative volume <= 6 Angstrom^3) start end length 1 23 26 4 2 85 91 7 3 95 95 1 4 99 99 1 5 102 105 4 6 122 133 12 7 174 176 3 8 182 191 10write.pdb(xyz=pdb$xyz[inds$xyz],resno=pdb$atom[inds$atom,"resno"], file="core.pdb") ##- Fit trj onto starting structure based on core indices xyz <- fit.xyz( fixed = pdb$xyz, mobile = trj, fixed.inds = inds$xyz, mobile.inds = inds$xyz) ##write.pdb(pdb=pdb, xyz=xyz, file="new_trj.pdb") ##write.ncdf(xyz, "new_trj.nc")